4CO6
 
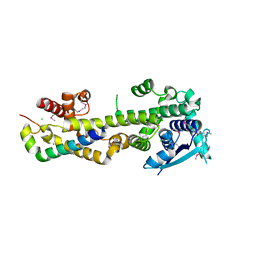 | | Crystal structure of the Nipah virus RNA free nucleoprotein- phosphoprotein complex | | Descriptor: | BROMIDE ION, CHLORIDE ION, NUCLEOPROTEIN, ... | | Authors: | Yabukarksi, F, Lawrence, P, Tarbouriech, N, Bourhis, J.M, Jensen, M.R, Ruigrok, R.W.H, Blackledge, M, Volchkov, V, Jamin, M. | | Deposit date: | 2014-01-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structure of Nipah Virus Unassembled Nucleoprotein in Complex with its Viral Chaperone.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2XD5
 
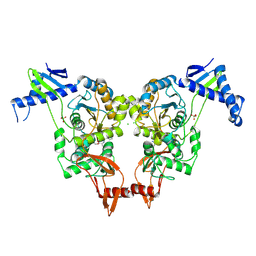 | | Structural insights into the catalytic mechanism and the role of Streptococcus pneumoniae PBP1b | | Descriptor: | CHLORIDE ION, N-BENZOYL-D-ALANINE, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, Lemaire, D, Jamin, M, Dideberg, O, Dessen, A. | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism and the Role of Streptococcus Pneumoniae Pbp1B
To be Published
|
|
8FFR
 
 | | Revised structure of the rabies virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (99-MER) | | Authors: | Leyrat, C, Bourhis, J.M, Albertini, A.A.V, Wernimont, A.K, Muziol, T, Ravelli, R.B.G, Weissenhorn, W, Ruigrok, R.W.H, Jamin, M. | | Deposit date: | 2022-12-09 | | Release date: | 2023-01-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure and Dynamics of the Unassembled Nucleoprotein of Rabies Virus in Complex with Its Phosphoprotein Chaperone Module.
Viruses, 14, 2022
|
|
8B8V
 
 | |
8BJW
 
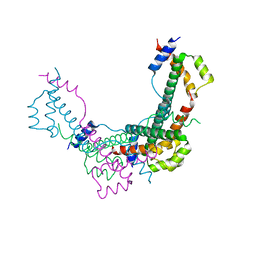 | |
2K47
 
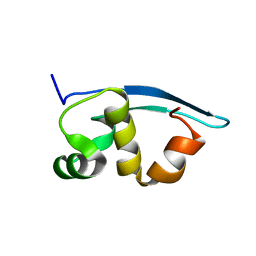 | | Solution structure of the C-terminal N-RNA binding domain of the Vesicular Stomatitis Virus Phosphoprotein | | Descriptor: | Phosphoprotein | | Authors: | Ribeiro, E.A, Favier, A, Gerard, F.C, Leyrat, C, Brutscher, B, Blondel, D, Ruigrok, R.W, Blackledge, M, Jamin, M. | | Deposit date: | 2008-05-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-Terminal Nucleoprotein-RNA Binding Domain of the Vesicular Stomatitis Virus Phosphoprotein.
J.Mol.Biol., 2008
|
|
7PON
 
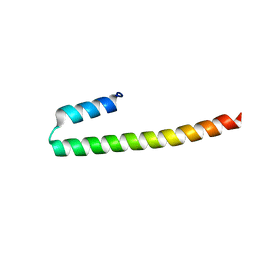 | |
7PNO
 
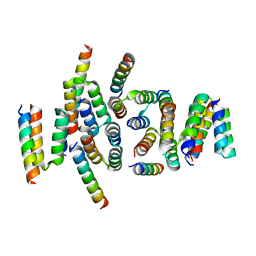 | | C terminal domain of Nipah Virus Phosphoprotein fused to the Ntail alpha more of the Nucleoprotein. | | Descriptor: | Phosphoprotein, alpha MoRE of Nipah virus Nucleoprotein tail | | Authors: | Bourhis, J.M, Yabukaski, F, Tarbouriech, N, Jamin, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Dynamics of the C-terminal X Domain of Nipah and Hendra Viruses Controls the Attachment to the C-terminal Tail of the Nucleocapsid Protein.
J.Mol.Biol., 434, 2022
|
|
3PMK
 
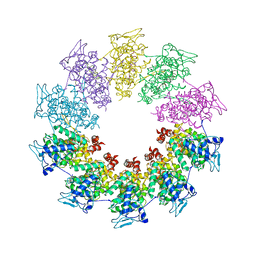 | | Crystal structure of the Vesicular Stomatitis Virus RNA free nucleoprotein/phosphoprotein complex | | Descriptor: | Nucleocapsid protein, Phosphoprotein | | Authors: | Leyrat, C, Yabukarski, F, Tarbouriech, N, Ruigrok, R.W.H, Jamin, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure of the Vesicular Stomatitis Virus N0-P Complex
Plos Pathog., 7, 2011
|
|
3T4R
 
 | |
4HEO
 
 | |
2WFH
 
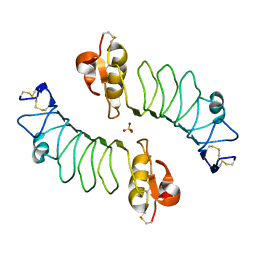 | | The Human Slit 2 Dimerization Domain D4 | | Descriptor: | SLIT HOMOLOG 2 PROTEIN C-PRODUCT, SULFATE ION | | Authors: | Seiradake, E, von Philipsborn, A.C, Henry, M, Fritz, M, Lortat-Jacob, H, Jamin, M, Hemrika, W, Bastmeyer, M, Cusack, S, McCarthy, A.A. | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and functional relevance of the Slit2 homodimerization domain.
EMBO Rep., 10, 2009
|
|
3L32
 
 | |
4GJW
 
 | |
4UDE
 
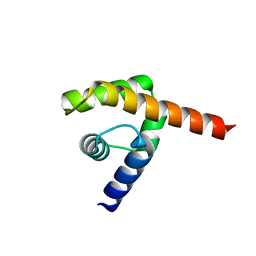 | | An oligomerization domain confers pioneer properties to the LEAFY master floral regulator | | Descriptor: | GINLFY PROTEIN, GLYCEROL, TETRAETHYLENE GLYCOL | | Authors: | Nanao, M.H, Sayou, C, Dumas, R, Parcy, F. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Sam Oligomerization Domain Shapes the Genomic Binding Landscape of the Leafy Transcription Factor
Nat.Commun., 7, 2016
|
|
4A1F
 
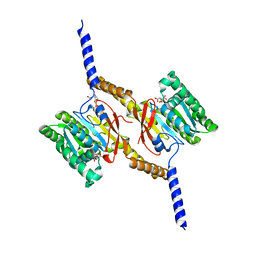 | | Crystal structure of C-terminal domain of Helicobacter pylori DnaB Helicase | | Descriptor: | CITRATE ANION, REPLICATIVE DNA HELICASE | | Authors: | Stelter, M, Kapp, U, Timmins, J, Terradot, L. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of a Dodecameric Bacterial Replicative Helicase.
Structure, 20, 2012
|
|
3K90
 
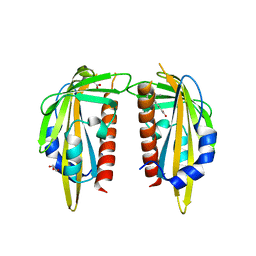 | | The Abscisic acid receptor PYR1 in complex with Abscisic Acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Dupeux, F.D, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2009-10-15 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The abscisic acid receptor PYR1 in complex with abscisic acid.
Nature, 462, 2009
|
|
4OJC
 
 | |
4OJE
 
 | |
4OJD
 
 | |
2XZE
 
 | |
