2D25
 
 | | C-C-A-G-G-C-M5C-T-G-G; HELICAL FINE STRUCTURE, HYDRATION, AND COMPARISON WITH C-C-A-G-G-C-C-T-G-G | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*(5CM)P*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Heinemann, U, Hahn, M. | | Deposit date: | 1991-04-23 | | Release date: | 1991-04-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G.
J.Biol.Chem., 267, 1992
|
|
9DNA
 
 | |
1D26
 
 | |
1BD1
 
 | | CRYSTALLOGRAPHIC STUDY OF ONE TURN OF G/C-RICH B-DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G)-3'), TRIETHYLAMMONIUM ION | | Authors: | Heinemann, U. | | Deposit date: | 1989-08-16 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic study of one turn of G/C-rich B-DNA.
J.Mol.Biol., 210, 1989
|
|
1CGC
 
 | |
3ANA
 
 | |
6HD3
 
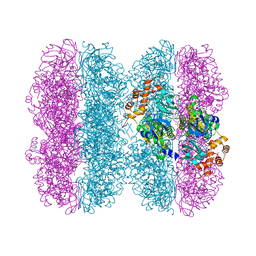 | | Common mode of remodeling AAA ATPases p97/CDC48 by their disassembly cofactors ASPL/PUX1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48 homolog A, PHOSPHATE ION | | Authors: | Heinemann, U, Roske, Y, Banchenko, S, Arumughan, A, Petrovic, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Common Mode of Remodeling AAA ATPases p97/CDC48 by Their Disassembling Cofactors ASPL/PUX1.
Structure, 27, 2019
|
|
6HD0
 
 | |
9RNT
 
 | |
2AAD
 
 | | THE ROLE OF HISTIDINE-40 IN RIBONUCLEASE T1 CATALYSIS: THREE-DIMENSIONAL STRUCTURES OF THE PARTIALLY ACTIVE HIS40LYS MUTANT | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 ISOZYME | | Authors: | Zegers, I, Verhelst, P, Choe, C.W, Steyaert, J, Heinemann, U, Wyns, L, Saenger, W. | | Deposit date: | 1992-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of histidine-40 in ribonuclease T1 catalysis: three-dimensionalstructures of the partially active His40Lys mutant.
Biochemistry, 31, 1992
|
|
2AAE
 
 | | THE ROLE OF HISTIDINE-40 IN RIBONUCLEASE T1 CATALYSIS: THREE-DIMENSIONAL STRUCTURES OF THE PARTIALLY ACTIVE HIS40LYS MUTANT | | Descriptor: | CALCIUM ION, PHOSPHATE ION, RIBONUCLEASE T1 | | Authors: | Zegers, I, Verhelst, P, Choe, C.W, Steyaert, J, Heinemann, U, Wyns, L, Saenger, W. | | Deposit date: | 1992-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of histidine-40 in ribonuclease T1 catalysis: three-dimensionalstructures of the partially active His40Lys mutant.
Biochemistry, 31, 1992
|
|
3RIQ
 
 | | Siphovirus 9NA tailspike receptor binding domain | | Descriptor: | GLYCEROL, Tailspike protein | | Authors: | Andres, D, Roske, Y, Doering, C, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2011-04-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tail morphology controls DNA release in two Salmonella phages with one lipopolysaccharide receptor recognition system.
Mol.Microbiol., 83, 2012
|
|
3JZ7
 
 | |
3RNT
 
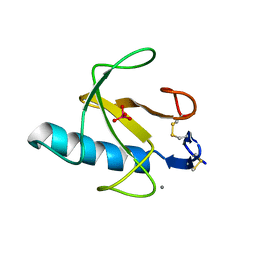 | | CRYSTAL STRUCTURE OF GUANOSINE-FREE RIBONUCLEASE T1, COMPLEXED WITH VANADATE(V), SUGGESTS CONFORMATIONAL CHANGE UPON SUBSTRATE BINDING | | Descriptor: | CALCIUM ION, RIBONUCLEASE T1, VANADATE ION | | Authors: | Kostrewa, D, Choe, H.-W, Heinemann, U, Saenger, W. | | Deposit date: | 1989-05-31 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of guanosine-free ribonuclease T1, complexed with vanadate (V), suggests conformational change upon substrate binding.
Biochemistry, 28, 1989
|
|
1QYM
 
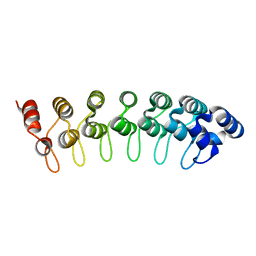 | | X-ray structure of human gankyrin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10 | | Authors: | Manjasetty, B.A, Quedenau, C, Sievert, V, Buessow, K, Niesen, F, Delbrueck, H, Heinemann, U. | | Deposit date: | 2003-09-11 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of human gankyrin, the product of a gene linked to hepatocellular carcinoma.
Proteins, 55, 2004
|
|
1RNT
 
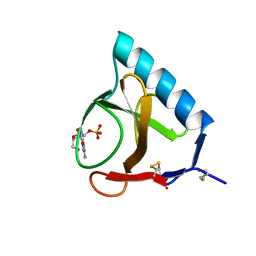 | | RESTRAINED LEAST-SQUARES REFINEMENT OF THE CRYSTAL STRUCTURE OF THE RIBONUCLEASE T1(ASTERISK)2(PRIME)-GUANYLIC ACID COMPLEX AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 ISOZYME | | Authors: | Saenger, W, Arni, R, Heinemann, U, Tokuoka, R. | | Deposit date: | 1987-07-10 | | Release date: | 1987-10-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restrained Least-Squares Refinement of the Crystal Structure of the Ribonuclease T1(Asterisk)2(Prime)-Guanylic Acid Complex at 1.9 Angstroms Resolution
Acta Crystallogr.,Sect.B, 43, 1987
|
|
1GPP
 
 | |
5MR7
 
 | |
5MPH
 
 | | Structural Basis of Gene Regulation by the Grainyhead Transcription Factor Superfamily | | Descriptor: | Grainyhead-like protein 1 homolog, IODIDE ION | | Authors: | Ming, Q, Roske, Y, Schuetz, A, Walentin, K, Ibraimi, I, Schmidt-Ott, K.M, Heinemann, U. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.337 Å) | | Cite: | Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family.
Nucleic Acids Res., 46, 2018
|
|
4XNF
 
 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XOP
 
 | | Tailspike protein double mutant D339N/E372Q of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XMY
 
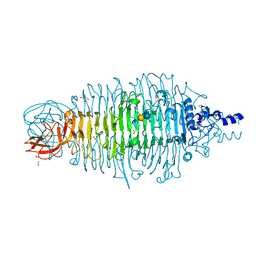 | | Tailspike protein double mutant D339A/E372A of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4URR
 
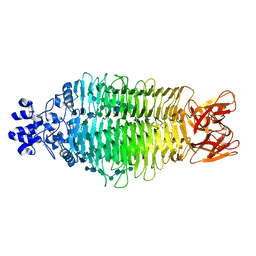 | | Tailspike protein of Sf6 bacteriophage bound to Shigella flexneri O- antigen octasaccharide fragment | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL TAIL PROTEIN, MANGANESE (II) ION, ... | | Authors: | Gohlke, U, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2014-07-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bacteriophage Tailspikes and Bacterial O-Antigens as a Model System to Study Weak-Affinity Protein-Polysaccharide Interactions.
J.Am.Chem.Soc., 138, 2016
|
|
1LRA
 
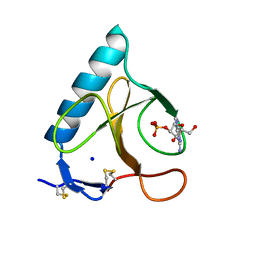 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
122D
 
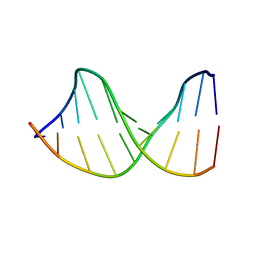 | |
