6IL7
 
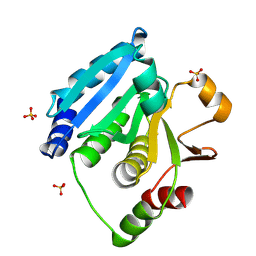 | | Structure of Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit F (AhpF) C503A mutant | | Descriptor: | SULFATE ION, Thioredoxin reductase/glutathione-related protein | | Authors: | Toh, Y.K, Shin, J, Balakrishna, A.M, Eisenhaber, F, Eisenhaber, B, Gruber, G. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effect of the additional cysteine 503 of vancomycin-resistant Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit F (AhpF) and the mechanism of AhpF and subunit C assembling.
Free Radic. Biol. Med., 138, 2019
|
|
5X09
 
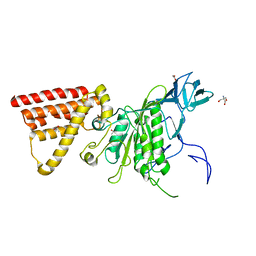 | | Crystal structure of subunit A mutant P235A/S238C of the A-ATP synthase from pyrococcus horikoshii OT3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Dhirendra, S, Gruber, G. | | Deposit date: | 2017-01-20 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic and enzymatic insights into the mechanisms of Mg-ADP inhibition in the A1 complex of the A1AO ATP synthase
J. Struct. Biol., 201, 2018
|
|
1D8S
 
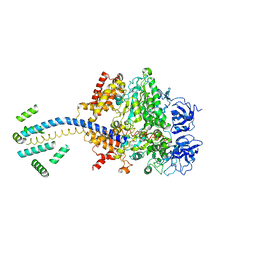 | | ESCHERICHIA COLI F1 ATPASE | | Descriptor: | F1 ATPASE (ALPHA SUBUNIT), F1 ATPASE (BETA SUBUNIT), F1 ATPASE (GAMMA SUBUNIT) | | Authors: | Hausrath, A.C, Gruber, G, Matthews, B.W, Capaldi, R.A. | | Deposit date: | 1999-10-25 | | Release date: | 1999-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structural features of the gamma subunit of the Escherichia coli F(1) ATPase revealed by a 4.4-A resolution map obtained by x-ray crystallography.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3TIV
 
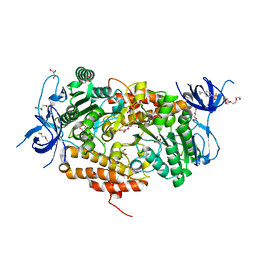 | | Crystal structure of subunit B mutant N157A of the A1AO ATP synthase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-08-22 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
3SSA
 
 | | Crystal structure of subunit B mutant N157T of the A1AO ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sundararaman, L, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-07-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
3TGW
 
 | | Crystal structure of subunit B mutant H156A of the A1AO ATP synthase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-08-18 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
3M4Y
 
 | | Structural characterization of the subunit A mutant P235A of the A-ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Manimekalai, M.S, Balakrishna, A.M, Kumar, A, Priya, R, Biukovic, G, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-03-12 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
8HCR
 
 | |
2K6I
 
 | | The domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H1-47 | | Descriptor: | Uncharacterized protein MJ0223 | | Authors: | Biukovic, N, Gayen, S, Pervushin, K, Gruber, G, Biukovic, G. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H(1-47).
Biophys.J., 97, 2009
|
|
2KHK
 
 | | NMR solution structure of the b30-82 domain of subunit b of Escherichia coli F1FO ATP synthase | | Descriptor: | ATP synthase subunit b | | Authors: | Priya, R, Biukovic, G, Gayen, S, Vivekanandan, S, Gruber, G. | | Deposit date: | 2009-04-08 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure, determined by nuclear magnetic resonance, of the b30-82 domain of subunit b of Escherichia coli F1Fo ATP synthase
J.Bacteriol., 191, 2009
|
|
2RKW
 
 | | Intermediate position of ATP on its trail to the binding pocket inside the subunit B mutant R416W of the energy converter A1Ao ATP synthase | | Descriptor: | V-type ATP synthase beta chain | | Authors: | Kumar, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Gruber, G. | | Deposit date: | 2007-10-18 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Spectroscopic and crystallographic studies of the mutant R416W give insight into the nucleotide binding traits of subunit B of the A1Ao ATP synthase
Proteins, 75, 2009
|
|
3B2Q
 
 | | Intermediate position of ATP on its trail to the binding pocket inside the subunit B mutant R416W of the energy converter A1Ao ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, ... | | Authors: | Kumar, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Gruber, G. | | Deposit date: | 2007-10-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Spectroscopic and crystallographic studies of the mutant R416W give insight into the nucleotide binding traits of subunit B of the A1Ao ATP synthase
Proteins, 75, 2009
|
|
3P20
 
 | | Crystal structure of vanadate bound subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-10-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
3QG1
 
 | | Crystal structure of P-loop G239A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, V-type ATP synthase alpha chain | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-24 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3QJY
 
 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3QIA
 
 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3MFY
 
 | | Structural characterization of the subunit A mutant F236A of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Balakrishna, A.M, Kumar, A, Manimekali, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-04-05 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
2KWY
 
 | | Structure of G61-101 | | Descriptor: | V-type proton ATPase subunit G | | Authors: | Rishikesan, S, Gruber, G. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of G61-101
To be Published
|
|
2KZ9
 
 | |
2KPA
 
 | | Specific motifs of the V-ATPase a2-subunit isoform interact with catalytic and regulatory domains of ARNO | | Descriptor: | ARNO(375-400) | | Authors: | Merkulova, M, Bakulina, A, Thaker, Y.R, Gruber, G, Marshansky, V. | | Deposit date: | 2009-10-11 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Specific motifs of the V-ATPase a2-subunit isoform interact with catalytic and regulatory domains of ARNO
Biochim.Biophys.Acta, 2010
|
|
3HGF
 
 | | Expression, purification, spectroscopical and crystallographical studies of segments of the nucleotide binding domain of the reticulocyte binding protein Py235 of Plasmodium yoelii | | Descriptor: | Rhoptry protein fragment | | Authors: | Gruber, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Jeyakanthan, J, Preiser, P.R, Gruber, G. | | Deposit date: | 2009-05-13 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural determination of functional units of the nucleotide binding domain (NBD94) of the reticulocyte binding protein Py235 of Plasmodium yoelii
Plos One, 5, 2010
|
|
6LXG
 
 | | NMR solution structure of regulatory ACT domain of the Mycobacterium tuberculosis Rel protein | | Descriptor: | GTP pyrophosphokinase | | Authors: | Shin, J, Singal, B, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2020-02-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of, and valine binding to the regulatory ACT domain of the Mycobacterium tuberculosis Rel protein.
Febs J., 288, 2021
|
|
2KK7
 
 | |
2K88
 
 | | Association of subunit d (Vma6p) and E (Vma4p) with G (Vma10p) and the NMR solution structure of subunit G (G1-59) of the Saccharomyces cerevisiae V1VO ATPase | | Descriptor: | Vacuolar proton pump subunit G | | Authors: | Sankaranarayanan, N, Gayen, S, Thaker, Y, Subramanian, V, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2008-09-04 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Assembly of subunit d (Vma6p) and G (Vma10p) and the NMR solution structure of subunit G (G(1-59)) of the Saccharomyces cerevisiae V(1)V(O) ATPase.
Biochim.Biophys.Acta, 1787, 2009
|
|
4QL9
 
 | |
