5U6G
 
 | |
3CR6
 
 | |
4LPC
 
 | |
3COP
 
 | |
3D96
 
 | |
8SDB
 
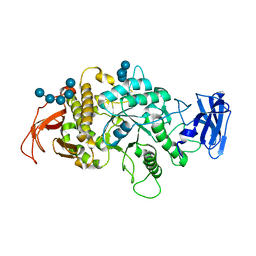 | | Crystal Structure of E.Coli Branching Enzyme in complex with malto-octose | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Bingham, C.R, Nayebi, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Maltooctaose-Bound Escherichia coli Branching Enzyme Suggests a Mechanism for Donor Chain Specificity.
Molecules, 28, 2023
|
|
4I9R
 
 | |
3F8A
 
 | | Crystal Structure of the R132K:R111L:L121E:R59W Mutant of Cellular Retinoic Acid-Binding Protein Type II Complexed with C15-aldehyde (a retinal analog) at 1.95 Angstrom resolution. | | Descriptor: | 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing Wavelength Regulation with an Engineered Rhodopsin Mimic and a C15-Retinal Analogue
Chempluschem, 77, 2012
|
|
3FA6
 
 | |
4I9S
 
 | |
3F9D
 
 | |
9O2O
 
 | |
9NG7
 
 | |
9O2N
 
 | |
9O2P
 
 | |
9NF1
 
 | |
9NG8
 
 | |
3I17
 
 | |
9D8M
 
 | |
9D8N
 
 | | Q108K:K40L:T51V:T53C:R58W:Y19W:Q38L:Q4A:T29L mutant of hCRBPII bound to FR1V and irradiated with UV light at pH 4 | | Descriptor: | (4aP)-N,N-diethyl-9,9-dimethyl-7-[(1E)-prop-1-en-1-yl]-9H-fluoren-2-amine, ACETIC ACID, GLYCEROL, ... | | Authors: | Bingham, C, Ehyaei, N, Geiger, J.H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Photoswitching fluorescence via a novel Trp-flap mechanism
To Be Published
|
|
9DDU
 
 | |
9DF1
 
 | |
8W02
 
 | |
8VZZ
 
 | | Q108K:K40L:T51V:T53S:Y19W:R58W mutant of hCRBPII bound to synthetic fluorophore TD-1V | | Descriptor: | (2E)-3-{5-[4-(dimethylamino)phenyl]thiophen-2-yl}but-2-enal, ACETATE ION, Retinol-binding protein 2 | | Authors: | Nosssoni, Z, Bingham, C.R, Geiger, J.H, Borhan, B. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Regulation of Absorption and Emission in a Protein/Fluorophore Complex.
Acs Chem.Biol., 19, 2024
|
|
8W00
 
 | |
