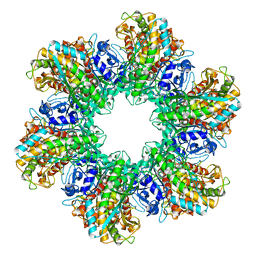
2GLS
 
 | |
2MLT
 
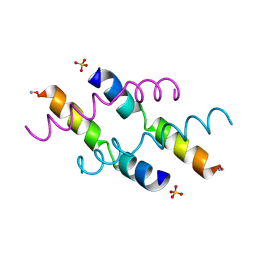 | |
2LGS
 
 | | FEEDBACK INHIBITION OF FULLY UNADENYLYLATED GLUTAMINE SYNTHETASE FROM SALMONELLA TYPHIMURIUM BY GLYCINE, ALANINE, AND SERINE | | Descriptor: | GLUTAMIC ACID, GLUTAMINE SYNTHETASE, MANGANESE (II) ION | | Authors: | Liaw, S.-H, Eisenberg, D. | | Deposit date: | 1994-08-05 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Feedback inhibition of fully unadenylylated glutamine synthetase from Salmonella typhimurium by glycine, alanine, and serine.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
6E78
 
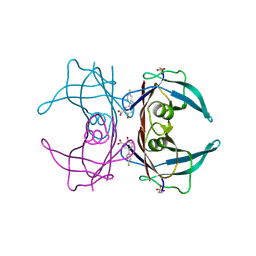 | | Structure of Human Transthyretin Asp38Ala Mutant in Complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
2ERL
 
 | | PHEROMONE ER-1 FROM | | Descriptor: | ETHANOL, MATING PHEROMONE ER-1 | | Authors: | Anderson, D.H, Weiss, M.S, Eisenberg, D. | | Deposit date: | 1995-12-20 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3AL1
 
 | | DESIGNED PEPTIDE ALPHA-1, RACEMIC P1BAR FORM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOLAMINE, PROTEIN (D, ... | | Authors: | Patterson, W.R, Anderson, D.H, Degrado, W.F, Cascio, D, Eisenberg, D. | | Deposit date: | 1998-10-26 | | Release date: | 1998-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Centrosymmetric bilayers in the 0.75 A resolution structure of a designed alpha-helical peptide, D,L-Alpha-1.
Protein Sci., 8, 1999
|
|
1BYZ
 
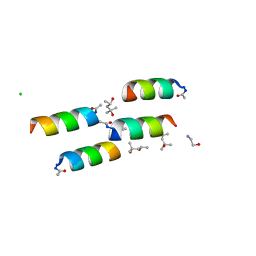 | | DESIGNED PEPTIDE ALPHA-1, P1 FORM | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Prive, G.G, Anderson, D.H, Wesson, L, Cascio, D, Eisenberg, D. | | Deposit date: | 1998-10-20 | | Release date: | 1998-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Packed protein bilayers in the 0.90 A resolution structure of a designed alpha helical bundle.
Protein Sci., 8, 1999
|
|
1SGK
 
 | | NUCLEOTIDE-FREE DIPHTHERIA TOXIN | | Descriptor: | DIPHTHERIA TOXIN (DIMERIC) | | Authors: | Bell, C.E, Eisenberg, D. | | Deposit date: | 1996-09-12 | | Release date: | 1996-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of nucleotide-free diphtheria toxin.
Biochemistry, 36, 1997
|
|
1RLC
 
 | | CRYSTAL STRUCTURE OF THE UNACTIVATED RIBULOSE 1, 5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE COMPLEXED WITH A TRANSITION STATE ANALOG, 2-CARBOXY-D-ARABINITOL 1,5-BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Cascio, D, Eisenberg, D. | | Deposit date: | 1993-08-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition state analog, 2-carboxy-D-arabinitol 1,5-bisphosphate.
Protein Sci., 3, 1994
|
|
1TOX
 
 | | DIPHTHERIA TOXIN DIMER COMPLEXED WITH NAD | | Descriptor: | DIPHTHERIA TOXIN (DIMERIC), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Bell, C.E, Eisenberg, D. | | Deposit date: | 1995-10-06 | | Release date: | 1996-06-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide.
Biochemistry, 35, 1996
|
|
1DTP
 
 | |
1RLD
 
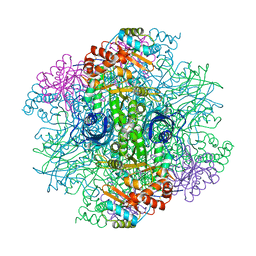 | | SOLID-STATE PHASE TRANSITION IN THE CRYSTAL STRUCTURE OF RIBULOSE 1,5-BIPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Eisenberg, D. | | Deposit date: | 1993-12-10 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solid-state phase transition in the crystal structure of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
2QZV
 
 | | Draft Crystal Structure of the Vault Shell at 9 Angstroms Resolution | | Descriptor: | Major vault protein | | Authors: | Anderson, D.H, Kickhoefer, V.A, Sievers, S.A, Rome, L.H, Eisenberg, D. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Draft crystal structure of the vault shell at 9-A resolution.
Plos Biol., 5, 2007
|
|
2APQ
 
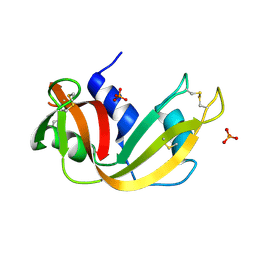 | | Crystal Structure of an Active Site Mutant of Bovine Pancreatic Ribonuclease A (H119A-RNase A) with a 10-Glutamine expansion in the C-terminal hinge-loop. | | Descriptor: | PHOSPHATE ION, Ribonuclease | | Authors: | Sambashivan, S, Liu, Y, Sawaya, M.R, Gingery, M, Eisenberg, D. | | Deposit date: | 2005-08-16 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure.
Nature, 437, 2005
|
|
2JCW
 
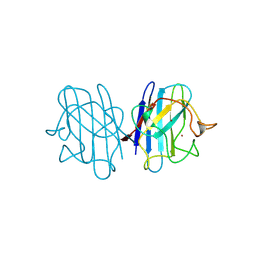 | | REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (I) ION, CU/ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-21 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
2Y3L
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 2 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3K
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 1 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3J
 
 | | Structure of segment AIIGLM from the amyloid-beta peptide (Ab, residues 30-35) | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y29
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph III | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y2A
 
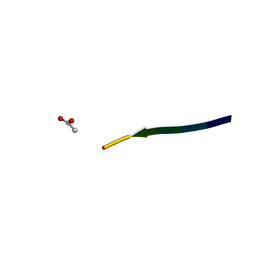 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph I | | Descriptor: | ACETATE ION, AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3L1F
 
 | | Bovine AlphaA crystallin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin A chain | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2009-12-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
3L1E
 
 | | Bovine AlphaA crystallin Zinc Bound | | Descriptor: | Alpha-crystallin A chain, GLYCEROL, ZINC ION | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2009-12-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
3L1G
 
 | | Human AlphaB crystallin | | Descriptor: | Alpha-crystallin B chain, SULFATE ION | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2009-12-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
1B4T
 
 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1B4L
 
 | | 15 ATMOSPHERE OXYGEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
