1BR4
 
 | | SMOOTH MUSCLE MYOSIN MOTOR DOMAIN-ESSENTIAL LIGHT CHAIN COMPLEX WITH MGADP.BEF3 BOUND AT THE ACTIVE SITE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Dominguez, R, Trybus, K.M, Cohen, C. | | Deposit date: | 1998-08-27 | | Release date: | 1998-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state.
Cell(Cambridge,Mass.), 94, 1998
|
|
1CEN
 
 | |
1CEO
 
 | |
1BR2
 
 | | SMOOTH MUSCLE MYOSIN MOTOR DOMAIN COMPLEXED WITH MGADP.ALF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN, ... | | Authors: | Dominguez, R, Trybus, K.M, Cohen, C. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BR1
 
 | | SMOOTH MUSCLE MYOSIN MOTOR DOMAIN-ESSENTIAL LIGHT CHAIN COMPLEX WITH MGADP.ALF4 BOUND AT THE ACTIVE SITE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN, ... | | Authors: | Dominguez, R, Trybus, K.M, Cohen, C. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state.
Cell(Cambridge,Mass.), 94, 1998
|
|
1CEC
 
 | |
6PSD
 
 | |
6PSE
 
 | |
1XYZ
 
 | |
7JPN
 
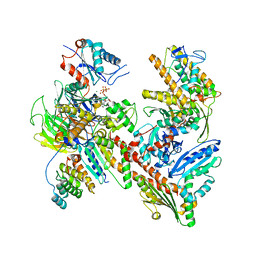 | | Cryo-EM structure of Arpin-bound Arp2/3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | van Eeuwen, T, Fregoso, F.E, Dominguez, R, Zimmet, A, Boczkowska, M, Rebowski, G. | | Deposit date: | 2020-08-09 | | Release date: | 2022-02-09 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Molecular mechanism of Arp2/3 complex inhibition by Arpin.
Nat Commun, 13, 2022
|
|
8TAH
 
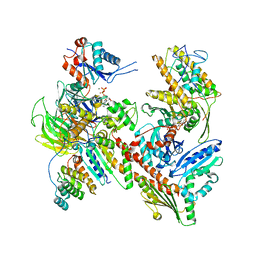 | | Cryo-EM structure of Cortactin-bound to Arp2/3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1A, ... | | Authors: | Fregoso, F.E, van Eeuwen, T, Dominguez, R. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanism of synergistic activation of Arp2/3 complex by cortactin and WASP-family proteins.
Nat Commun, 14, 2023
|
|
5V7X
 
 | | Crystal Structure of Myosin 1b residues 1-728 with bound sulfate and Calmodulin | | Descriptor: | Calmodulin-1, SULFATE ION, Unconventional myosin-Ib | | Authors: | Zwolak, A, Shuman, H, Dominguez, R, Ostap, E.M. | | Deposit date: | 2017-03-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.141 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1N2D
 
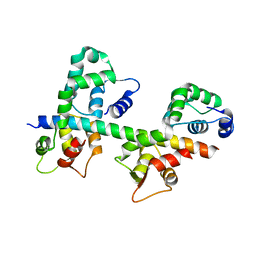 | | Ternary complex of MLC1P bound to IQ2 and IQ3 of Myo2p, a class V myosin | | Descriptor: | IQ2 AND IQ3 MOTIFS FROM MYO2P, A CLASS V MYOSIN, Myosin Light Chain | | Authors: | Terrak, M, Wu, G, Stafford, W.F, Lu, R.C, Dominguez, R. | | Deposit date: | 2002-10-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the light chain-binding domain of myosin V.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6NBW
 
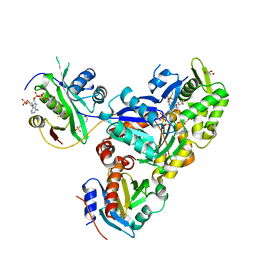 | | Ternary Complex of Beta/Gamma-Actin with Profilin and AnCoA-NAA80 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2018-12-10 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of actin N-terminal acetylation.
Sci Adv, 6, 2020
|
|
6NAS
 
 | | Ternary Complex of Ac-Alpha-Actin with Profilin and AcCoA-NAA80 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2018-12-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of actin N-terminal acetylation.
Sci Adv, 6, 2020
|
|
6NBE
 
 | | Ternary Complex of Ac-Alpha-Actin with Profilin and CoA-NAA80 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2018-12-07 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of actin N-terminal acetylation.
Sci Adv, 6, 2020
|
|
5TGC
 
 | | Structure of the hetero-trimer of Rtt102-Arp7/9 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-like protein ARP9, Actin-related protein 7, ... | | Authors: | Turegun, B, Dominguez, R. | | Deposit date: | 2016-09-27 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Actin-related proteins regulate the RSC chromatin remodeler by weakening intramolecular interactions of the Sth1 ATPase.
Commun Biol, 1, 2018
|
|
5WFN
 
 | | Revised model of leiomodin 2-mediated actin regulation (alternate refinement of PDB 4RWT) | | Descriptor: | Actin-5C, Leiomodin-2, MAGNESIUM ION, ... | | Authors: | Yurtsever, Z, Eck, M.J, Dominguez, R. | | Deposit date: | 2017-07-12 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of leiomodin 2 in complex with actin: a structural and functional reexamination
Biophys.J., 113, 2017
|
|
6UHC
 
 | | CryoEM structure of human Arp2/3 complex with bound NPFs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | Zimmet, A, van Eeuwen, T, Dominguez, R. | | Deposit date: | 2019-09-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism.
Sci Adv, 6, 2020
|
|
1NWK
 
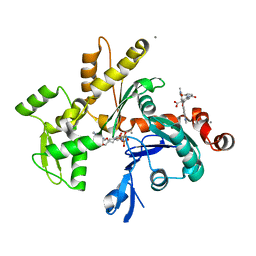 | | CRYSTAL STRUCTURE OF MONOMERIC ACTIN IN THE ATP STATE | | Descriptor: | Actin, alpha skeletal muscle, CALCIUM ION, ... | | Authors: | Graceffa, P, Dominguez, R. | | Deposit date: | 2003-02-06 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics.
J.Biol.Chem., 278, 2003
|
|
1S70
 
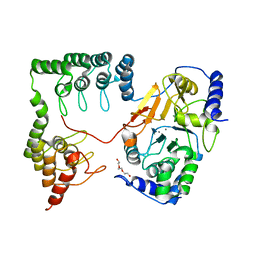 | | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) | | Descriptor: | 130 kDa myosin-binding subunit of smooth muscle myosin phophatase (M130), MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-beta (or delta) catalytic subunit, ... | | Authors: | Kerff, F, Terrak, M, Dominguez, R. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of protein phosphatase 1 regulation
Nature, 429, 2004
|
|
7T5Q
 
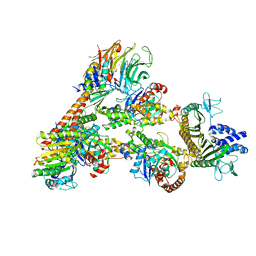 | | Cryo-EM Structure of a Transition State of Arp2/3 Complex Activation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Rebowski, G, van Eeuwen, T, Boczkowska, M, Dominguez, R. | | Deposit date: | 2021-12-13 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of an Arp2/3 complex mini-branch capped by capping protein (CapZ)
To Be Published
|
|
8F8Q
 
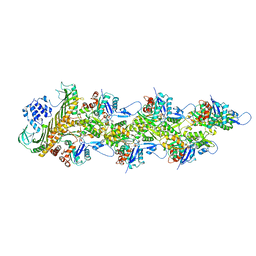 | | Cryo-EM structure of the CapZ-capped barbed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
8F8R
 
 | | Cryo-EM structure of the free barbed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
8F8T
 
 | | Cryo-EM structure of the Tropomodulin-capped pointed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
