8BNA
 
 | | BINDING OF HOECHST 33258 TO THE MINOR GROOVE OF B-DNA | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Pjura, P.E, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1986-08-29 | | Release date: | 1987-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of Hoechst 33258 to the minor groove of B-DNA.
J.Mol.Biol., 197, 1987
|
|
5BNA
 
 | | THE PRIMARY MODE OF BINDING OF CISPLATIN TO A B-DNA DODECAMER: C-G-C-G-A-A-T-T-C-G-C-G | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM TRIAMINE ION | | Authors: | Wing, R.M, Pjura, P, Drew, H.R, Dickerson, R.E. | | Deposit date: | 1983-08-22 | | Release date: | 1983-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The primary mode of binding of cisplatin to a B-DNA dodecamer: C-G-C-G-A-A-T-T-C-G-C-G
EMBO J., 3, 1984
|
|
1HCR
 
 | |
4BNA
 
 | |
3BNA
 
 | |
6BNA
 
 | | BINDING OF AN ANTITUMOR DRUG TO DNA. NETROPSIN AND C-G-C-G-A-A-T-T-BRC-G-C-G | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(CBR)P*GP*CP*G)-3'), NETROPSIN | | Authors: | Kopka, M.L, Yoon, C, Goodsell, D, Pjura, P, Dickerson, R.E. | | Deposit date: | 1984-08-30 | | Release date: | 1984-10-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G.
J.Mol.Biol., 183, 1985
|
|
2BNA
 
 | |
7BNA
 
 | |
451C
 
 | |
351C
 
 | |
1BNA
 
 | | STRUCTURE OF A B-DNA DODECAMER. CONFORMATION AND DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Drew, H.R, Wing, R.M, Takano, T, Broka, C, Tanaka, S, Itakura, K, Dickerson, R.E. | | Deposit date: | 1981-01-26 | | Release date: | 1981-05-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a B-DNA dodecamer: conformation and dynamics.
Proc.Natl.Acad.Sci.USA, 78, 1981
|
|
3DNB
 
 | |
1ZG5
 
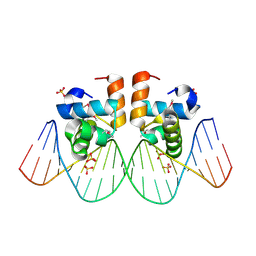 | | NarL complexed to narG-89 promoter palindromic tail-to-tail DNA site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*CP*TP*AP*TP*AP*GP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/nitrite response regulator protein narL, SULFATE ION | | Authors: | Maris, A.E, Kaczor-Grzeskowiak, M, Ma, Z, Kopka, M.L, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2005-04-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Biochemistry, 44, 2005
|
|
1ZG1
 
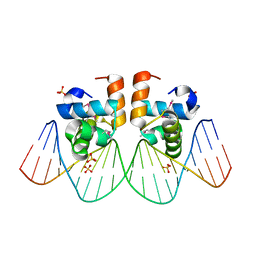 | | NarL complexed to nirB promoter non-palindromic tail-to-tail DNA site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*GP*GP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*CP*TP*CP*CP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/nitrite response regulator protein narL, ... | | Authors: | Maris, A.E, Kaczor-Grzeskowiak, M, Ma, Z, Kopka, M.L, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2005-04-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Biochemistry, 44, 2005
|
|
1ANA
 
 | |
5DNB
 
 | |
4FIS
 
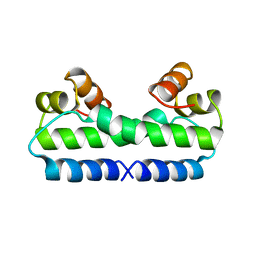 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J.-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1ZNA
 
 | |
334D
 
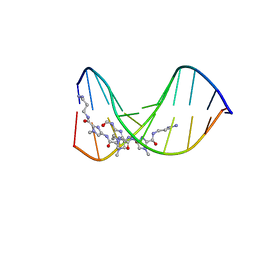 | |
307D
 
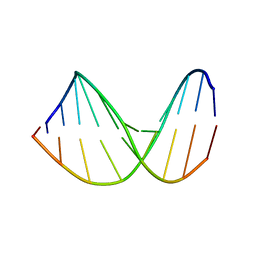 | | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3') | | Authors: | Han, G.W, Kopka, M.L, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1997-01-07 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis.
J.Mol.Biol., 269, 1997
|
|
455D
 
 | |
101D
 
 | |
1A04
 
 | | THE STRUCTURE OF THE NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL IN THE MONOCLINIC C2 CRYSTAL FORM | | Descriptor: | NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL | | Authors: | Baikalov, I, Schroder, I, Kaczor-Grzeskowiak, M, Cascio, D, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NarL dimerization? Suggestive evidence from a new crystal form
Biochemistry, 37, 1998
|
|
3FIS
 
 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
274D
 
 | | CRYSTAL STRUCTURE OF A COVALENT DNA-DRUG ADDUCT: ANTHRAMYCIN BOUND TO C-C-A-A-C-G-T-T-G-G, AND A MOLECULAR EXPLANATION OF SPECIFICITY | | Descriptor: | ANTHRAMYCIN, DNA (5'-D(*CP*CP*AP*AP*CP*GP*TP*TP*(DRUG)GP*G)-3') | | Authors: | Kopka, M.L, Goodsell, D.S, Baikalov, I, Grzeskowiak, K, Cascio, D, Dickerson, R.E. | | Deposit date: | 1994-04-28 | | Release date: | 1994-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a covalent DNA-drug adduct: anthramycin bound to C-C-A-A-C-G-T-T-G-G and a molecular explanation of specificity.
Biochemistry, 33, 1994
|
|
