1NUH
 
 | |
1GZD
 
 | | Crystal structure of pig phosphoglucose isomerase | | Descriptor: | GLUCOSE-6-PHOSPHATE ISOMERASE, SULFATE ION | | Authors: | Davies, C, Muirhead, H. | | Deposit date: | 2002-05-20 | | Release date: | 2002-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Phosphoglucose Isomerase from Pig Muscle and its Complex with 5-Phosphoarabinonate
Proteins, 49, 2002
|
|
1GZV
 
 | |
1HD8
 
 | |
1SEI
 
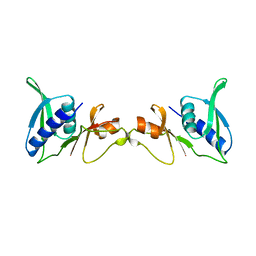 | | STRUCTURE OF 30S RIBOSOMAL PROTEIN S8 | | Descriptor: | RIBOSOMAL PROTEIN S8 | | Authors: | Davies, C, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-08-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence for specific S8-RNA and S8-protein interactions within the 30S ribosomal subunit: ribosomal protein S8 from Bacillus stearothermophilus at 1.9 A resolution.
Structure, 4, 1996
|
|
1NLD
 
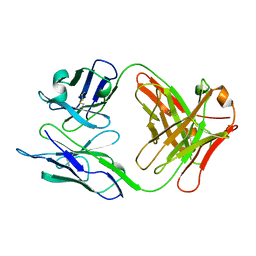 | | FAB FRAGMENT OF A NEUTRALIZING ANTIBODY DIRECTED AGAINST AN EPITOPE OF GP41 FROM HIV-1 | | Descriptor: | FAB1583 | | Authors: | Davies, C, Beauchamp, J.C, Emery, D, Rawas, A, Muirhead, H. | | Deposit date: | 1996-07-02 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Fab fragment from a neutralizing monoclonal antibody directed against an epitope of gp41 from HIV-1.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1N8T
 
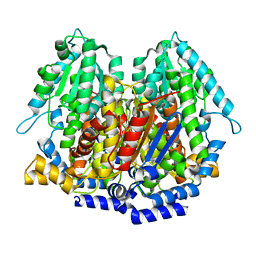 | |
1WHI
 
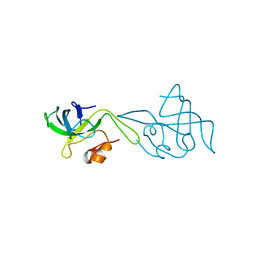 | | RIBOSOMAL PROTEIN L14 | | Descriptor: | RIBOSOMAL PROTEIN L14 | | Authors: | Davies, C, White, S.W, Ramakrishnan, V. | | Deposit date: | 1996-01-10 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of ribosomal protein L14 reveals an important organizational component of the translational apparatus.
Structure, 4, 1996
|
|
1EBL
 
 | | THE 1.8 A CRYSTAL STRUCTURE AND ACTIVE SITE ARCHITECTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE III (FABH) FROM ESCHERICHIA COLI | | Descriptor: | BETA-KETOACYL-ACP SYNTHASE III, COENZYME A | | Authors: | Davies, C, Heath, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2000-01-24 | | Release date: | 2000-02-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from escherichia coli.
Structure Fold.Des., 8, 2000
|
|
3UPN
 
 | |
3UPP
 
 | |
3UPO
 
 | |
3QTG
 
 | |
7MIE
 
 | | Crystal structure of the Borreliella burgdorferi PlzA protein in complex with c-di-GMP | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, ... | | Authors: | Davies, C, Singh, A. | | Deposit date: | 2021-04-16 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution crystal structure of the Borreliella burgdorferi PlzA protein in complex with c-di-GMP: new insights into the interaction of c-di-GMP with the novel xPilZ domain.
Pathog Dis, 79, 2021
|
|
2OFS
 
 | | Crystal structure of human CD59 | | Descriptor: | CD59 glycoprotein | | Authors: | Davies, C. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CD59: implications for molecular recognition of the complement proteins C8 and C9 in the membrane-attack complex.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G5D
 
 | |
4U3T
 
 | |
9MD0
 
 | | Crystal structure of the transpeptidase domain of PBP2 from the Neisseria gonorrhoeae cephalosporin decreased susceptibility strain 35/02 in complex with boronate inhibitor VNRX-6752 | | Descriptor: | (3R)-3-({(2R)-2-(4-carboxyphenyl)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]acetyl}amino)-2-hydroxy-3,4-dihydro-2H-1,2-benzoxaborinine-8-carboxylic acid, Penicillin-binding protein 2 | | Authors: | Stratton, C.M, Bala, S, Davies, C. | | Deposit date: | 2024-12-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | A new class of penicillin-binding protein inhibitors to address drug-resistant Neisseria gonorrhoeae.
Biorxiv, 2024
|
|
9MCZ
 
 | | Crystal structure of the transpeptidase domain of PBP2 from the Neisseria gonorrhoeae cephalosporin decreased susceptibility strain 35/02 in complex with boronate inhibitor VNRX-6884 | | Descriptor: | (3R)-3-{[(2R)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]-2-(4-phosphonophenyl)acetyl]amino}-2-hydroxy-3,4-dihydro-2H-1,2-benzoxaborinine-8-carboxylic acid, Penicillin-binding protein 2 | | Authors: | Stratton, C.M, Bala, S, Davies, C. | | Deposit date: | 2024-12-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A new class of penicillin-binding protein inhibitors to address drug-resistant Neisseria gonorrhoeae.
Biorxiv, 2024
|
|
1TZC
 
 | | Crystal structure of phosphoglucose/phosphomannose isomerase from Pyrobaculum aerophilum in complex with 5-phosphoarabinonate | | Descriptor: | 5-PHOSPHOARABINONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Swan, M.K, Hansen, T, Schoenheit, P, Davies, C. | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A novel phosphoglucose/phosphomannose isomease from the crenarchaeon Pyrobaculum aerophilum is a member of the PGI superfamily: structural evidence at 1.16 A resolution
J.Biol.Chem., 279, 2004
|
|
1TZB
 
 | | Crystal structure of native phosphoglucose/phosphomannose isomerase from Pyrobaculum aerophilum | | Descriptor: | GLYCEROL, SULFATE ION, glucose-6-phosphate isomerase, ... | | Authors: | Swan, M.K, Hansen, T, Schoenheit, P, Davies, C. | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A novel phosphoglucose/phosphomannose isomease from the crenarchaeon Pyrobaculum aerophilum is a member of the PGI superfamily: structural evidence at 1.16 A resolution
J.Biol.Chem., 279, 2004
|
|
1U0F
 
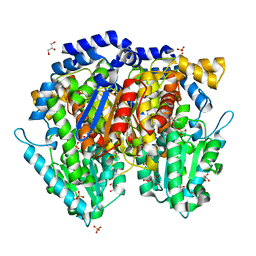 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0E
 
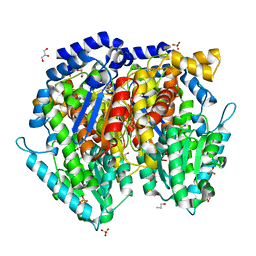 | | Crystal structure of mouse phosphoglucose isomerase | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0G
 
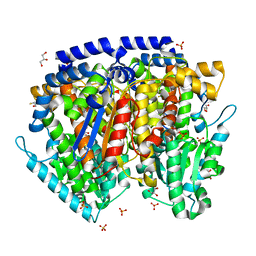 | | Crystal structure of mouse phosphoglucose isomerase in complex with erythrose 4-phosphate | | Descriptor: | BETA-MERCAPTOETHANOL, ERYTHOSE-4-PHOSPHATE, GLYCEROL, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
8VBZ
 
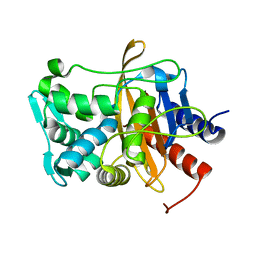 | |
