2Y24
 
 | | STRUCTURAL BASIS FOR SUBSTRATE RECOGNITION BY ERWINIA CHRYSANTHEMI GH5 GLUCURONOXYLANASE | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, IMIDAZOLE, TETRAETHYLENE GLYCOL, ... | | Authors: | Urbanikova, L, Vrsanska, M, Krogh, K.B, Hoff, T, Biely, P. | | Deposit date: | 2010-12-13 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Basis for Substrate Recognition by Erwinia Chrysanthemi Gh30 Glucuronoxylanase.
FEBS J., 278, 2011
|
|
3VNZ
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with D-glucuronic acid | | Descriptor: | GLYCEROL, PHOSPHATE ION, beta-D-glucopyranuronic acid, ... | | Authors: | Momma, M, Fujimoto, Z, Michikawa, M, Ichinose, H, Yoshida, M, Kotake, Y, Biely, P, Tsumuraya, Y, Kaneko, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical characterization of glycoside hydrolase family 79 beta-glucuronidase from Acidobacterium capsulatum
J.Biol.Chem., 287, 2012
|
|
3VNY
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum | | Descriptor: | GLYCEROL, PHOSPHATE ION, beta-GLUCURONIDASE | | Authors: | Momma, M, Fujimoto, Z, Michikawa, M, Ichinose, H, Yoshida, M, Kotake, Y, Biely, P, Tsumuraya, Y, Kaneko, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical characterization of glycoside hydrolase family 79 beta-glucuronidase from Acidobacterium capsulatum
J.Biol.Chem., 287, 2012
|
|
3VO0
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum covalent-bonded with 2-deoxy-2-fluoro-D-glucuronic acid | | Descriptor: | 2,4-DINITROPHENOL, 2-deoxy-2-fluoro-alpha-D-glucopyranuronic acid, 2-deoxy-2-fluoro-beta-D-glucopyranuronic acid, ... | | Authors: | Momma, M, Fujimoto, Z, Michikawa, M, Ichinose, H, Jongkees, S, Yoshida, M, Kotake, Y, Biely, P, Tsumuraya, Y, Withers, S, Kaneko, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of glycoside hydrolase family 79 beta-glucuronidase from Acidobacterium capsulatum
J.Biol.Chem., 287, 2012
|
|
2CC0
 
 | | Family 4 carbohydrate esterase from Streptomyces lividans in complex with acetate | | Descriptor: | ACETATE ION, ACETYL-XYLAN ESTERASE, ZINC ION | | Authors: | Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Vincent, F, Brzozowski, A.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Puchart, V, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases
J.Biol.Chem., 281, 2006
|
|
2C71
 
 | | The structure of a family 4 acetyl xylan esterase from Clostridium thermocellum in complex with a magnesium ion. | | Descriptor: | GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, DOCKERIN TYPE I:POLYSACCHARIDE, ... | | Authors: | Taylor, E.J, Turkenburg, P.J, Vincent, F, Brzozowski, A.M, Gloster, T.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2005-11-17 | | Release date: | 2006-01-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell-Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases.
J.Biol.Chem., 281, 2006
|
|
2C79
 
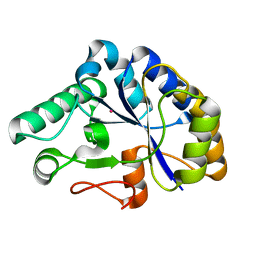 | | The structure of a family 4 acetyl xylan esterase from Clostridium thermocellum in complex with a colbalt ion. | | Descriptor: | COBALT (II) ION, GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, ... | | Authors: | Taylor, E.J, Turkenburg, P.J, Vincent, F, Brzozowski, A.M, Gloster, T.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2005-11-18 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases.
J.Biol.Chem., 281, 2006
|
|
1VLQ
 
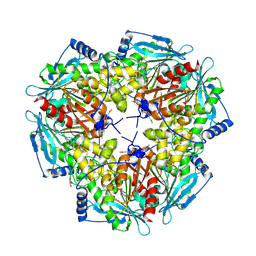 | |
1H41
 
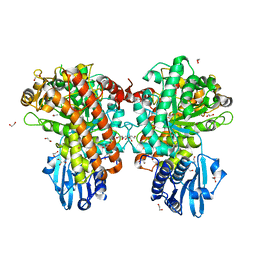 | | Pseudomonas cellulosa E292A alpha-D-glucuronidase mutant complexed with aldotriuronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid, ALPHA-GLUCURONIDASE, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-09-25 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Alpha-Glucuronidase,Glca67A,of Cellvibrio Japonicus Utilizes the Carboxylate and Methyl Groups of Aldobiouronic Acid as Important Substrate Recognition Determinants
J.Biol.Chem., 278, 2003
|
|
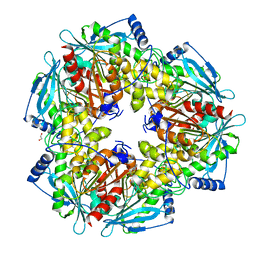
3M82
 
 | |
3M81
 
 | |
3M83
 
 | |
3FVR
 
 | |
3FYT
 
 | |
3FVT
 
 | |
3FYU
 
 | |
3PIC
 
 | |
