8TIM
 
 | |
2LZH
 
 | | THE STRUCTURES OF THE MONOCLINIC AND ORTHORHOMBIC FORMS OF HEN EGG-WHITE LYSOZYME AT 6 ANGSTROMS RESOLUTION. | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Artymiuk, P.J, Blake, C.C.F, Rice, D.W, Wilson, K.S. | | Deposit date: | 1981-06-29 | | Release date: | 1981-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution.
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1LZ1
 
 | |
1LZH
 
 | | THE STRUCTURES OF THE MONOCLINIC AND ORTHORHOMBIC FORMS OF HEN EGG-WHITE LYSOZYME AT 6 ANGSTROMS RESOLUTION. | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Artymiuk, P.J, Blake, C.C.F, Rice, D.W, Wilson, K.S. | | Deposit date: | 1981-06-29 | | Release date: | 1981-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution.
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1FHA
 
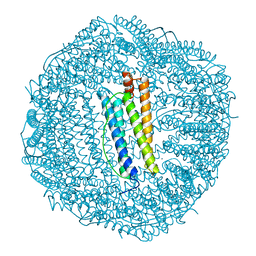 | |
1BDX
 
 | | E. COLI DNA HELICASE RUVA WITH BOUND DNA HOLLIDAY JUNCTION, ALPHA CARBONS AND PHOSPHATE ATOMS ONLY | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*GP*CP*AP*TP*AP*TP*GP*CP*AP*TP*GP*C)-3'), HOLLIDAY JUNCTION DNA HELICASE RUVA | | Authors: | Hargreaves, D, Rice, D.W, Sedelnikova, S.E, Artymiuk, P.J, Lloyd, R.G, Rafferty, J.B. | | Deposit date: | 1998-05-11 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal structure of E.coli RuvA with bound DNA Holliday junction at 6 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
2FHA
 
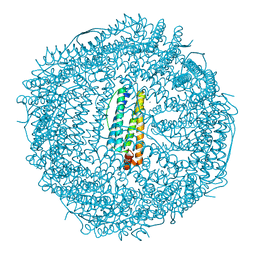 | | HUMAN H CHAIN FERRITIN | | Descriptor: | CALCIUM ION, FERRITIN | | Authors: | Hempstead, P.D, Artymiuk, P.J, Harrison, P.M. | | Deposit date: | 1997-03-03 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution.
J.Mol.Biol., 268, 1997
|
|
1AEW
 
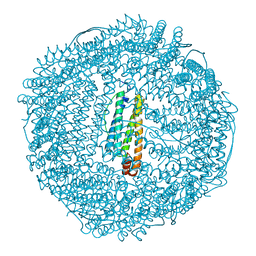 | | L-CHAIN HORSE APOFERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Hempstead, P.D, Yewdall, S.J, Lawson, D.M, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution.
J.Mol.Biol., 268, 1997
|
|
4K1P
 
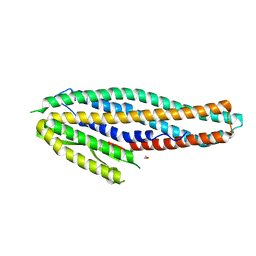 | | Structure of the NheA component of the Nhe toxin from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, NheA, SULFATE ION | | Authors: | Ganash, M, Phung, D, Artymiuk, P.J. | | Deposit date: | 2013-04-05 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the NheA Component of the Nhe Toxin from Bacillus cereus: Implications for Function.
Plos One, 8, 2013
|
|
1SUL
 
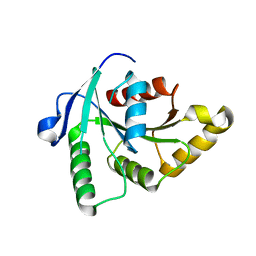 | | Crystal Structure of the apo-YsxC | | Descriptor: | GTP-binding protein YsxC | | Authors: | Ruzheinikov, S.N, Das, K.S, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
1SVI
 
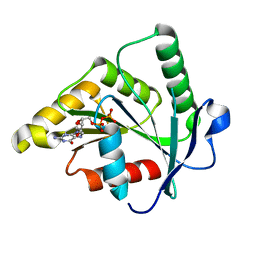 | | Crystal Structure of the GTP-binding protein YsxC complexed with GDP | | Descriptor: | GTP-binding protein YSXC, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
1SVW
 
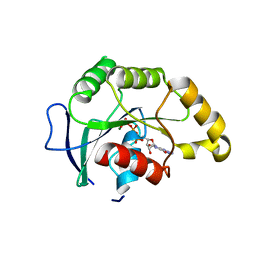 | | Crystal Structure of YsxC complexed with GMPPNP | | Descriptor: | GTP-binding protein YsxC, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-30 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
1ZUJ
 
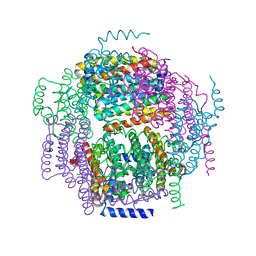 | | The crystal structure of the Lactococcus lactis MG1363 DpsA protein | | Descriptor: | hypothetical protein Llacc01001955 | | Authors: | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | Deposit date: | 2005-05-31 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
1ZS3
 
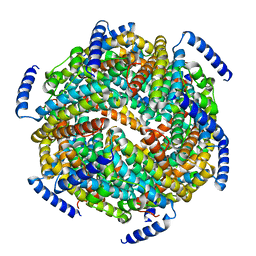 | | The crystal structure of the Lactococcus lactis MG1363 DpsB protein | | Descriptor: | Lactococcus lactis MG1363 DpsA | | Authors: | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | Deposit date: | 2005-05-23 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
3ZD9
 
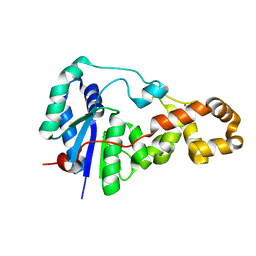 | | Potassium bound structure of E. coli ExoIX in P21 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
1HEW
 
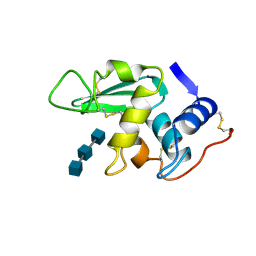 | |
1QOY
 
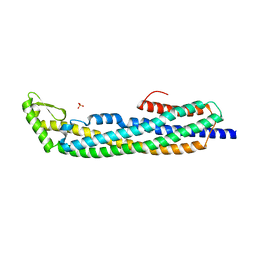 | | E.coli Hemolysin E (HlyE, ClyA, SheA) | | Descriptor: | HEMOLYSIN E, SULFATE ION | | Authors: | Wallace, A.J, Stillman, T.J, Atkins, A, Jamieson, S.J, Bullough, P.A, Green, J, Artymiuk, P.J. | | Deposit date: | 1999-11-25 | | Release date: | 2000-01-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | E. Coli Hemolysin E (Hlye, Clya, Shea): X-Ray Crystal Structure of the Toxin and Observation of Membrane Pores by Electron Microscopy
Cell(Cambridge,Mass.), 100, 2000
|
|
5HNK
 
 | | Crystal structure of T5Fen in complex intact substrate and metal ions. | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*C)-3'), Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Almalki, F.A, Feng, M, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HML
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Flemming, C.S, Feng, M, Sedelnikova, S.E, Zhang, J, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HMM
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exodeoxyribonuclease, ... | | Authors: | Flemming, C.S, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3TUA
 
 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) C94S mutant | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1) | | Authors: | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | Deposit date: | 2011-09-16 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
3TU8
 
 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | BROMIDE ION, Burkholderia Lethal Factor 1 (BLF1) | | Authors: | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | Deposit date: | 2011-09-16 | | Release date: | 2011-11-30 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
1EUM
 
 | | CRYSTAL STRUCTURE OF THE E.COLI FERRITIN ECFTNA | | Descriptor: | FERRITIN 1 | | Authors: | Stillman, T.J, Hempstead, P.D, Artymiuk, P.J, Andrews, S.C, Hudson, A.J, Treffry, A, Guest, J.R, Harrison, P.M. | | Deposit date: | 2000-04-17 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The high-resolution X-ray crystallographic structure of the ferritin (EcFtnA) of Escherichia coli; comparison with human H ferritin (HuHF) and the structures of the Fe(3+) and Zn(2+) derivatives.
J.Mol.Biol., 307, 2001
|
|
1JKU
 
 | | Crystal Structure of Manganese Catalase from Lactobacillus plantarum | | Descriptor: | CALCIUM ION, HYDROXIDE ION, MANGANESE (III) ION, ... | | Authors: | Barynin, V.V, Whittaker, M.M, Antonyuk, S.V, Lamzin, V.S, Harrison, P.M, Artymiuk, P.J, Whittaker, J.W. | | Deposit date: | 2001-07-13 | | Release date: | 2002-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of manganese catalase from Lactobacillus plantarum.
Structure, 9, 2001
|
|
1JKV
 
 | | Crystal Structure of Manganese Catalase from Lactobacillus plantarum complexed with azide | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CALCIUM ION, ... | | Authors: | Barynin, V.V, Whittaker, M.M, Antonyuk, S.V, Lamzin, V.S, Harrison, P.M, Artymiuk, P.J, Whittaker, J.W. | | Deposit date: | 2001-07-13 | | Release date: | 2002-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of manganese catalase from Lactobacillus plantarum.
Structure, 9, 2001
|
|
