2BKJ
 
 | |
1I8M
 
 | | CRYSTAL STRUCTURE OF A RECOMBINANT ANTI-SINGLE-STRANDED DNA ANTIBODY FRAGMENT COMPLEXED WITH DT5 | | Descriptor: | 5'-D(*TP*TP*TP*TP*T)-3', 5'-D(P*TP*T)-3', ANTIBODY HEAVY CHAIN FAB, ... | | Authors: | Tanner, J.J, Komissarov, A.A, Deutscher, S.L. | | Deposit date: | 2001-03-14 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of an Antigen-Binding Fragment Bound to Single-Stranded DNA
J.Mol.Biol., 314, 2001
|
|
1CER
 
 | |
1BKJ
 
 | | NADPH:FMN OXIDOREDUCTASE FROM VIBRIO HARVEYI | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-FLAVIN OXIDOREDUCTASE, PHOSPHATE ION | | Authors: | Tanner, J.J, Lei, B, TU, S.-C, Krause, K.L. | | Deposit date: | 1998-07-08 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flavin reductase P: structure of a dimeric enzyme that reduces flavin.
Biochemistry, 35, 1996
|
|
5JJG
 
 | | Structure of magnesium-loaded ALG-2 | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Pcalcium-binding protein ALG-2, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2016-04-23 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | EF5 Is the High-Affinity Mg(2+) Site in ALG-2.
Biochemistry, 55, 2016
|
|
1XKJ
 
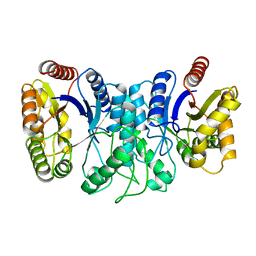 | | BACTERIAL LUCIFERASE BETA2 HOMODIMER | | Descriptor: | BETA2 LUCIFERASE | | Authors: | Tanner, J.J, Krause, K.L. | | Deposit date: | 1996-10-08 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial luciferase beta 2 homodimer: implications for flavin binding.
Biochemistry, 36, 1997
|
|
1XVJ
 
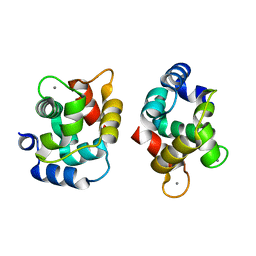 | | Crystal Structure Of Rat alpha-Parvalbumin D94S/G98E Mutant | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Tanner, J.J, Agah, S, Lee, Y.H, Henzl, M.T. | | Deposit date: | 2004-10-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the D94S/G98E Variant of Rat alpha-Parvalbumin. An Explanation for the Reduced Divalent Ion Affinity.
Biochemistry, 44, 2005
|
|
6X0K
 
 | |
5KF6
 
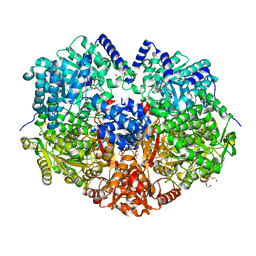 | |
6D97
 
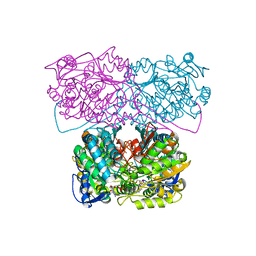 | |
5KOW
 
 | | Structure of rifampicin monooxygenase | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pentachlorophenol 4-monooxygenase | | Authors: | Tanner, J.J, Liu, L.-K. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase.
J.Biol.Chem., 291, 2016
|
|
4OE6
 
 | | Crystal Structure of Yeast ALDH4A1 | | Descriptor: | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | | Authors: | Tanner, J.J. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural Studies of Yeast Delta (1)-Pyrroline-5-carboxylate Dehydrogenase (ALDH4A1): Active Site Flexibility and Oligomeric State.
Biochemistry, 53, 2014
|
|
5VWT
 
 | |
5VWU
 
 | |
5KOX
 
 | |
8DKQ
 
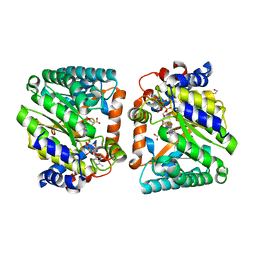 | |
8DKP
 
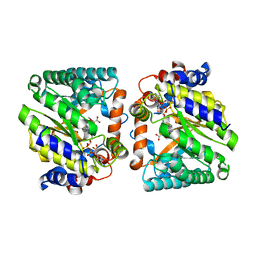 | |
8DKO
 
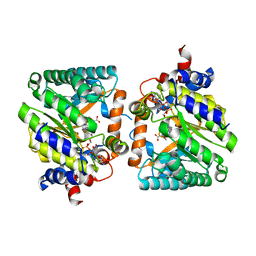 | |
6MVS
 
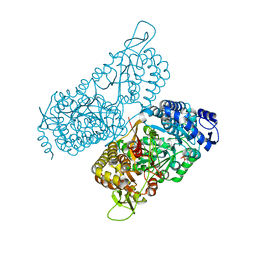 | | Structure of a bacterial ALDH16 complexed with NAD | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J, Liu, L. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
6MVT
 
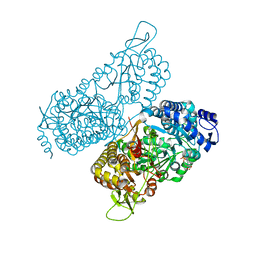 | | Structure of a bacterial ALDH16 complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Aldehyde dehydrogenase, SODIUM ION | | Authors: | Tanner, J.J, Liu, L. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
6MVR
 
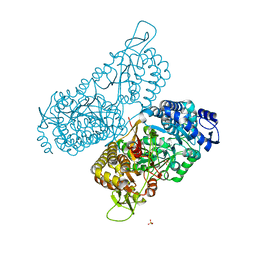 | | Structure of a bacterial ALDH16 | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Tanner, J.J, Liu, L. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
6MVU
 
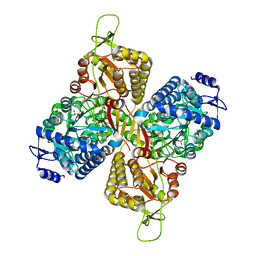 | |
3HAZ
 
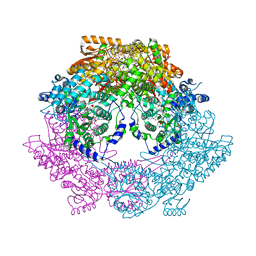 | | Crystal structure of bifunctional proline utilization A (PutA) protein | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-05-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ITG
 
 | |
7SQN
 
 | |
