4B8C
 
 | | nuclease module of the yeast Ccr4-Not complex | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, GLUCOSE-REPRESSIBLE ALCOHOL DEHYDROGENASE TRANSCRIPTIONAL EFFECTOR, POLY(A) RIBONUCLEASE POP2 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B8A
 
 | | Structure of yeast NOT1 MIF4G domain co-crystallized with CAF1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, POLY(A) RIBONUCLEASE POP2 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B89
 
 | | MIF4G domain of the yeast Not1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B8B
 
 | | N-Terminal domain of the yeast Not1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, GOLD ION | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
8C0T
 
 | | NRS 1.2: Fluorescent Sensors for Imaging Interstitial Calcium | | Descriptor: | CALCIUM ION, SULFATE ION, mNeonGreen,Optimized Ratiometric Calcium Sensor | | Authors: | Basquin, J, Griesbeck, O, Valiente-Gabioud, A. | | Deposit date: | 2022-12-19 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fluorescent sensors for imaging of interstitial calcium.
Nat Commun, 14, 2023
|
|
4CT6
 
 | | CNOT9-CNOT1 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
7OGG
 
 | | Nse5/6 complex | | Descriptor: | DNA repair protein KRE29,DNA repair protein KRE29,DNA repair protein KRE29, Non-structural maintenance of chromosome element 5,Non-structural maintenance of chromosome element 5,Non-structural maintenance of chromosome element 5 | | Authors: | Basquin, J, Taschner, M, Gruber, S. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Nse5/6 inhibits the Smc5/6 ATPase and modulates DNA substrate binding.
Embo J., 40, 2021
|
|
2YHT
 
 | |
2Y90
 
 | |
8BFI
 
 | | human CNOT1-CNOT10-CNOT11 module | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
8BFH
 
 | | CNOT11 | | Descriptor: | CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
8BFJ
 
 | | CNOT11-GGNBP2 complex | | Descriptor: | 1,2-ETHANEDIOL, CCR4-NOT transcription complex subunit 11, Gametogenetin-binding protein 2 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
7OCZ
 
 | |
7OCX
 
 | |
6YVH
 
 | | CWC22-CWC27-EIF4A3 Complex | | Descriptor: | Eukaryotic initiation factor 4A-III, Pre-mRNA-splicing factor CWC22 homolog, Spliceosome-associated protein CWC27 homolog | | Authors: | Basquin, J, Busetto, V, LeHir, H, Conti, E. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and functional insights into CWC27/CWC22 heterodimer linking the exon junction complex to spliceosomes.
Nucleic Acids Res., 48, 2020
|
|
5ANR
 
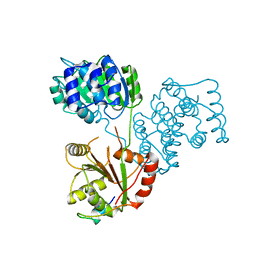 | | Structure of a human 4E-T - DDX6 - CNOT1 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TRANSPORTER, PROBABLE ATP-DEPENDENT RNA HELICASE DDX6 | | Authors: | Basquin, J, Oezguer, S, Conti, E. | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of a Human 4E-T/Ddx6/Cnot1 Complex Reveals the Different Interplay of Ddx6-Binding Proteins with the Ccr4-not Complex.
Cell Rep., 13, 2015
|
|
2WP8
 
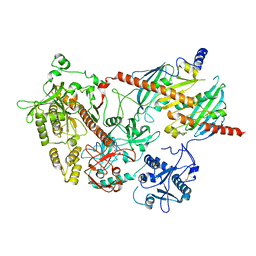 | | yeast rrp44 nuclease | | Descriptor: | CHLORIDE ION, EXOSOME COMPLEX COMPONENT RRP45, EXOSOME COMPLEX COMPONENT SKI6, ... | | Authors: | Basquin, J, Bonneau, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Yeast Exosome Functions as a Macromolecular Cage to Channel RNA Substrates for Degradation.
Cell(Cambridge,Mass.), 139, 2009
|
|
4CT7
 
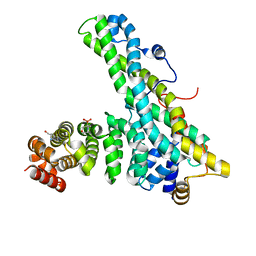 | | CNOT9-CNOT1 complex with bound tryptophan | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG, ... | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
4CV5
 
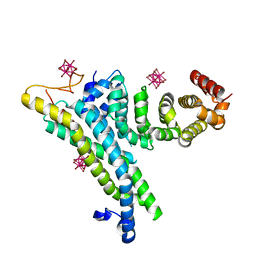 | | yeast NOT1 CN9BD-CAF40 complex | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, HEXATANTALUM DODECABROMIDE, PROTEIN CAF40 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2014-03-23 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.807 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
6I59
 
 | | Long wavelength native-SAD phasing of Sen1 helicase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
7AQO
 
 | | yeast THO-Sub2 complex dimer | | Descriptor: | BJ4_G0025130.mRNA.1.CDS.1, EM14S01-3B_G0007820.mRNA.1.CDS.1, TEX1 isoform 1, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
7APX
 
 | | yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2,Tho2, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
4CT5
 
 | | DDX6 | | Descriptor: | ACETATE ION, DDX6 | | Authors: | Ozgur, S, Basquin, J, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
6I5C
 
 | | Long wavelength native-SAD phasing of Tubulin-Stathmin-TTL complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
8RXB
 
 | | Human UPF1 CH domain in complex with SMG6 peptide | | Descriptor: | Regulator of nonsense transcripts 1, Telomerase-binding protein EST1A, ZINC ION | | Authors: | Langer, L, Basquin, J, Conti, E. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UPF1 helicase orchestrates mutually exclusive interactions with the SMG6 endonuclease and UPF2.
Nucleic Acids Res., 52, 2024
|
|
