2D25
 
 | | C-C-A-G-G-C-M5C-T-G-G; HELICAL FINE STRUCTURE, HYDRATION, AND COMPARISON WITH C-C-A-G-G-C-C-T-G-G | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*(5CM)P*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Heinemann, U, Hahn, M. | | Deposit date: | 1991-04-23 | | Release date: | 1991-04-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G.
J.Biol.Chem., 267, 1992
|
|
9DNA
 
 | |
1D26
 
 | |
1CGC
 
 | |
1BD1
 
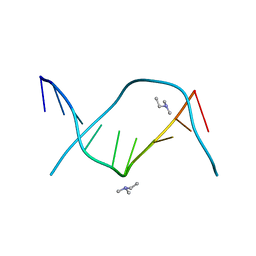 | | CRYSTALLOGRAPHIC STUDY OF ONE TURN OF G/C-RICH B-DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G)-3'), TRIETHYLAMMONIUM ION | | Authors: | Heinemann, U. | | Deposit date: | 1989-08-16 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic study of one turn of G/C-rich B-DNA.
J.Mol.Biol., 210, 1989
|
|
3ANA
 
 | |
6HD0
 
 | |
6HD3
 
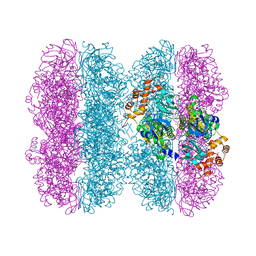 | | Common mode of remodeling AAA ATPases p97/CDC48 by their disassembly cofactors ASPL/PUX1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48 homolog A, PHOSPHATE ION | | Authors: | Heinemann, U, Roske, Y, Banchenko, S, Arumughan, A, Petrovic, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Common Mode of Remodeling AAA ATPases p97/CDC48 by Their Disassembling Cofactors ASPL/PUX1.
Structure, 27, 2019
|
|
2RNT
 
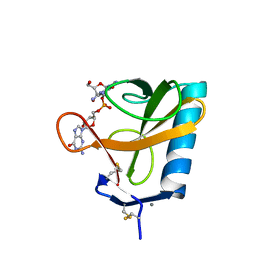 | | THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH GUANYLYL-2(PRIME),5(PRIME)-GUANOSINE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, GUANYLYL-2',5'-PHOSPHOGUANOSINE, RIBONUCLEASE T1 | | Authors: | Saenger, W, Koepke, J, Maslowska, M, Heinemann, U. | | Deposit date: | 1988-07-06 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of ribonuclease T1 complexed with guanylyl-2',5'-guanosine at 1.8 A resolution.
J.Mol.Biol., 206, 1989
|
|
9RNT
 
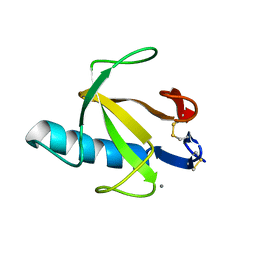 | |
4URR
 
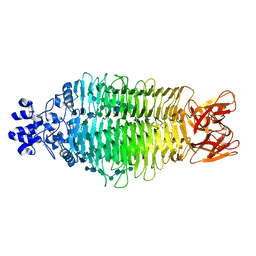 | | Tailspike protein of Sf6 bacteriophage bound to Shigella flexneri O- antigen octasaccharide fragment | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL TAIL PROTEIN, MANGANESE (II) ION, ... | | Authors: | Gohlke, U, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2014-07-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bacteriophage Tailspikes and Bacterial O-Antigens as a Model System to Study Weak-Affinity Protein-Polysaccharide Interactions.
J.Am.Chem.Soc., 138, 2016
|
|
3RNT
 
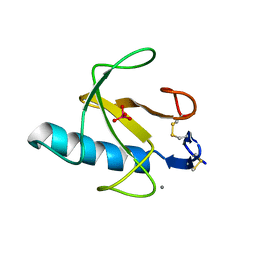 | | CRYSTAL STRUCTURE OF GUANOSINE-FREE RIBONUCLEASE T1, COMPLEXED WITH VANADATE(V), SUGGESTS CONFORMATIONAL CHANGE UPON SUBSTRATE BINDING | | Descriptor: | CALCIUM ION, RIBONUCLEASE T1, VANADATE ION | | Authors: | Kostrewa, D, Choe, H.-W, Heinemann, U, Saenger, W. | | Deposit date: | 1989-05-31 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of guanosine-free ribonuclease T1, complexed with vanadate (V), suggests conformational change upon substrate binding.
Biochemistry, 28, 1989
|
|
4ULW
 
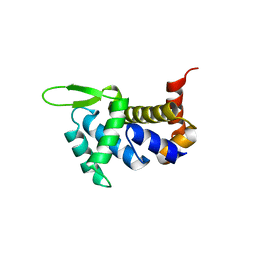 | |
4UUY
 
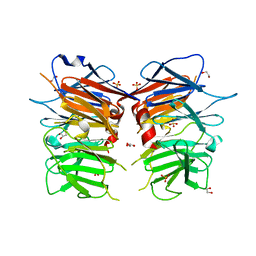 | | Structural Identification of the Vps18 beta-propeller reveals a critical role in the HOPS complex stability and function. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Behrmann, H, Gohlke, U, Heinemann, U. | | Deposit date: | 2014-08-01 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Identification of the Vps18 Beta-Propeller Reveals a Critical Role in the Hops Complex Stability and Function.
J.Biol.Chem., 289, 2014
|
|
2BT6
 
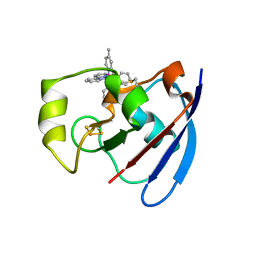 | | Ru(bpy)2(mbpy)-Modified Bovine Adrenodoxin | | Descriptor: | (4'-METHYL-2,2'BIPYRIDINE)BIS(2,2'-BIPYRIDINE), ADRENODOXIN 1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Halavaty, A, Mueller, J.J, Contzen, J, Jung, C, Hannemann, F, Bernhardt, R, Galander, M, Lendzian, F, Heinemann, U. | | Deposit date: | 2005-05-26 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Light-Induced Reduction of Bovine Adrenodoxin Via the Covalently Bound Ruthenium(II) Bipyridyl Complex: Intramolecular Electron Transfer and Crystal Structure.
Biochemistry, 45, 2006
|
|
5IFW
 
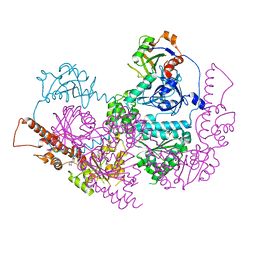 | |
5OF1
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 2-HYDROXYBENZOIC ACID, Spore coat-associated protein N, ethane-1,2-diol | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OF2
 
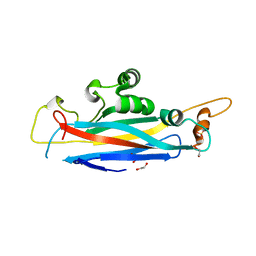 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Spore coat-associated protein N | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1RNT
 
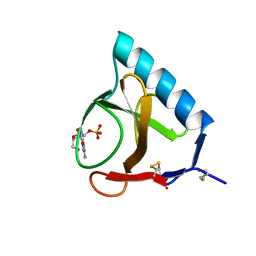 | | RESTRAINED LEAST-SQUARES REFINEMENT OF THE CRYSTAL STRUCTURE OF THE RIBONUCLEASE T1(ASTERISK)2(PRIME)-GUANYLIC ACID COMPLEX AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 ISOZYME | | Authors: | Saenger, W, Arni, R, Heinemann, U, Tokuoka, R. | | Deposit date: | 1987-07-10 | | Release date: | 1987-10-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restrained Least-Squares Refinement of the Crystal Structure of the Ribonuclease T1(Asterisk)2(Prime)-Guanylic Acid Complex at 1.9 Angstroms Resolution
Acta Crystallogr.,Sect.B, 43, 1987
|
|
2R80
 
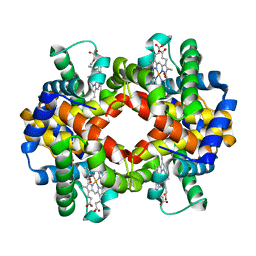 | | Pigeon Hemoglobin (OXY form) | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Ponnuswamy, M.N, Packianathan, C, Sundaresan, S, Neelagandan, K, Palani, K, Muller, J.J, Heinemann, U. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | X-ray crystal structure analysis of Hemolgobin from Pigeon (Columba Livia) at 1.44 angstrom
To be Published
|
|
1LRA
 
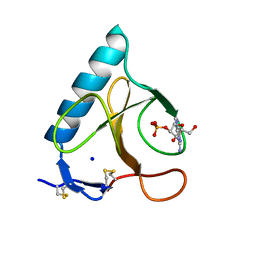 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
5JDT
 
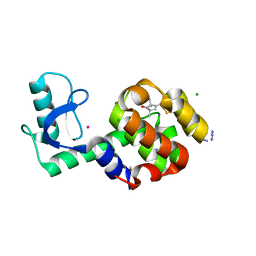 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at 100K | | Descriptor: | AZIDE ION, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2016-04-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
5NXT
 
 | | Wobble base paired RNA double helix | | Descriptor: | RNA (5'-R(*UP*GP*UP*UP*CP*UP*CP*UP*AP*CP*GP*AP*AP*GP*AP*AP*CP*A)-3') | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | A novel form of RNA double helix based on G·U and C·A+ wobble base pairing.
RNA, 24, 2018
|
|
5G27
 
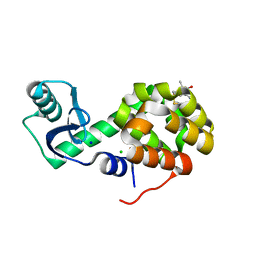 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at Room Temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ENDOLYSIN, ... | | Authors: | Gohlke, U, Consentius, P, Loll, B, Mueller, R, Kaupp, M, Heinemann, U, Risse, T. | | Deposit date: | 2016-04-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-Ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
3JZ7
 
 | |
