1QD7
 
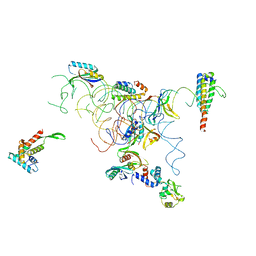 | | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | | Descriptor: | CENTRAL FRAGMENT OF 16 S RNA, END FRAGMENT OF 16 S RNA, S15 RIBOSOMAL PROTEIN, ... | | Authors: | Clemons Jr, W.M, May, J.L.C, Wimberly, B.T, McCutcheon, J.P, Capel, M.S, Ramakrishnan, V. | | Deposit date: | 1999-07-09 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution.
Nature, 400, 1999
|
|
1QB2
 
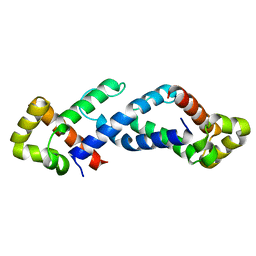 | | CRYSTAL STRUCTURE OF THE CONSERVED SUBDOMAIN OF HUMAN PROTEIN SRP54M AT 2.1A RESOLUTION: EVIDENCE FOR THE MECHANISM OF SIGNAL PEPTIDE BINDING | | Descriptor: | HUMAN SIGNAL RECOGNITION PARTICLE 54 KD PROTEIN | | Authors: | Clemons Jr, W.M, Gowda, K, Black, S.D, Zwieb, C, Ramakrishnan, V. | | Deposit date: | 1999-04-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 A resolution: evidence for the mechanism of signal peptide binding.
J.Mol.Biol., 292, 1999
|
|
7SPY
 
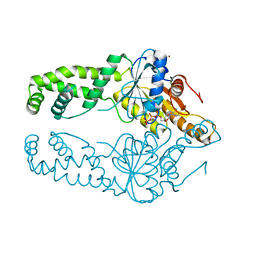 | | Get3 bound to ATP from G. intestinalis in the closed form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase ASNA1 homolog, MAGNESIUM ION, ... | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SQ0
 
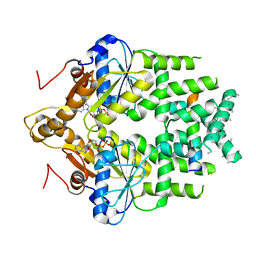 | | Get3 bound to ADP and the transmembrane domain of the tail-anchored protein Bos1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase ASNA1 homolog, MAGNESIUM ION, ... | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SPZ
 
 | | Nucleotide-free Get3 in two open forms | | Descriptor: | ATPase ASNA1 homolog, ZINC ION | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8ELF
 
 | | Structure of Get3d, a homolog of Get3, from Arabidopsis thaliana | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Manu, M.S, Barlow, A.N, Clemons Jr, W.M, Ramasamy, S. | | Deposit date: | 2022-09-23 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.
J.Biol.Chem., 299, 2023
|
|
8EGK
 
 | | Re-refinement of Crystal Structure of NosGet3d, the All4481 protein from Nostoc sp. PCC 7120 | | Descriptor: | AZIDE ION, All4481 protein | | Authors: | Barlow, A.N, Manu, M.S, Ramasamy, S, Clemons Jr, W.M. | | Deposit date: | 2022-09-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.
J.Biol.Chem., 299, 2023
|
|
1FJG
 
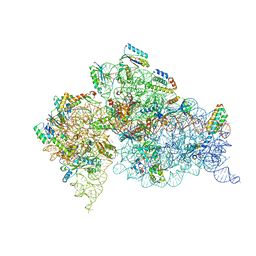 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Wimberly, B.T, Morgan-Warren, R.J, Ramakrishnan, V. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics
Nature, 407, 2000
|
|
1HNW
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH TETRACYCLINE | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HR0
 
 | | CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Morgan-Warren, R.J, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-20 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an initiation factor bound to the 30S ribosomal subunit.
Science, 291, 2001
|
|
1HNX
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH PACTAMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HNZ
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH HYGROMYCIN B | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1IBK
 
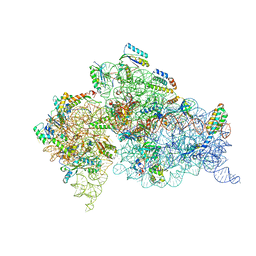 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTIC PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-04 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
1IBM
 
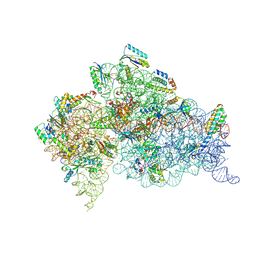 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-04 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
1IBL
 
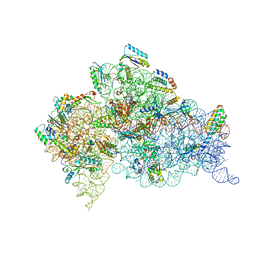 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE AND WITH THE ANTIBIOTIC PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-04 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
1J5E
 
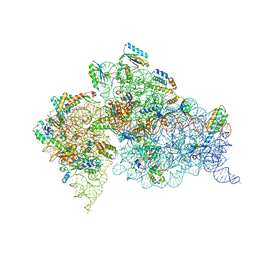 | | Structure of the Thermus thermophilus 30S Ribosomal Subunit | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Wimberly, B.T, Brodersen, D.E, Clemons Jr, W.M, Morgan-Warren, R, Carter, A.P, Vonrhein, C, Hartsch, T, Ramakrishnan, V. | | Deposit date: | 2002-04-08 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the 30S ribosomal subunit.
Nature, 407, 2000
|
|
1EMW
 
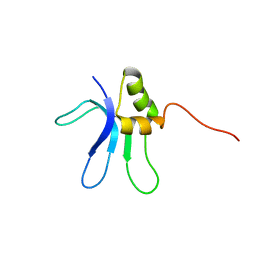 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S16 FROM THERMUS THERMOPHILUS | | Descriptor: | S16 RIBOSOMAL PROTEIN | | Authors: | Allard, P, Rak, A.V, Wimberly, B.T, Clemons Jr, W.M, Kalinin, A, Helgstrand, M, Garber, M.B, Ramakrishnan, V, Hard, T. | | Deposit date: | 2000-03-20 | | Release date: | 2000-08-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Another piece of the ribosome: solution structure of S16 and its location in the 30S subunit.
Structure Fold.Des., 8, 2000
|
|
1RH5
 
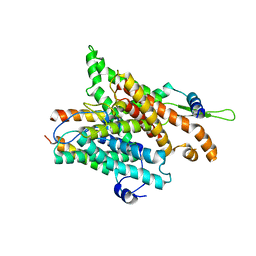 | | The structure of a protein conducting channel | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | Authors: | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a protein-conducting channel
Nature, 427, 2004
|
|
1TF2
 
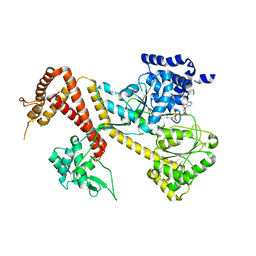 | |
1TF5
 
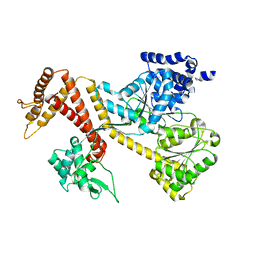 | |
1T1L
 
 | | Crystal structure of the long-chain fatty acid transporter FadL | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Long-chain fatty acid transport protein | | Authors: | van den Berg, B, Black, P.N, Clemons Jr, W.M, Rapoport, T.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the long-chain fatty acid transporter FadL.
Science, 304, 2004
|
|
1T16
 
 | | Crystal structure of the bacterial fatty acid transporter FadL from Escherichia coli | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, COPPER (II) ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | van den Berg, B, Black, P.N, Clemons Jr, W.M, Rapoport, T.A. | | Deposit date: | 2004-04-15 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the long-chain fatty acid transporter FadL.
Science, 304, 2004
|
|
1A32
 
 | |
4WWR
 
 | |
4PWX
 
 | |
