3KBR
 
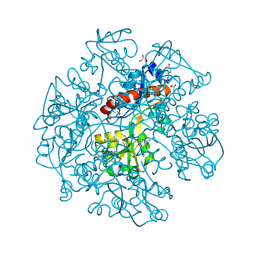 | | The crystal structure of cyclohexadienyl dehydratase precursor from Pseudomonas aeruginosa PA01 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cyclohexadienyl dehydratase, ... | | Authors: | Tan, K, Marshall, N, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | The crystal structure of cyclohexadienyl dehydratase precursor from Pseudomonas aeruginosa PA01
To be Published
|
|
3KKB
 
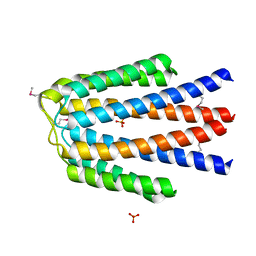 | |
3KD8
 
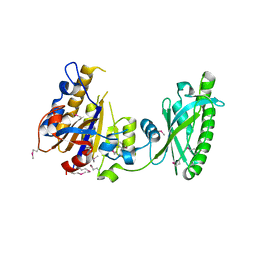 | |
3FF4
 
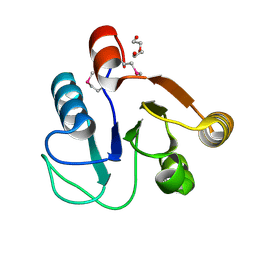 | |
3FNA
 
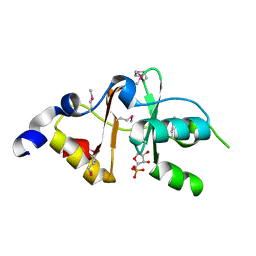 | |
3L34
 
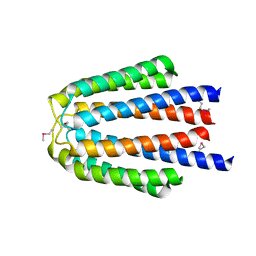 | |
3KYZ
 
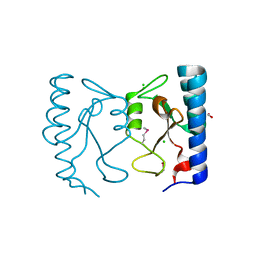 | | The crystal structure of the sensor domain of two-component sensor PfeS from Pseudomonas aeruginosa PA01 | | Descriptor: | CHLORIDE ION, FORMIC ACID, Sensor protein pfeS | | Authors: | Tan, K, Marshall, N, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-07 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | The crystal structure of the sensor domain of two-component sensor PfeS from Pseudomonas aeruginosa PA01
To be Published
|
|
3LDU
 
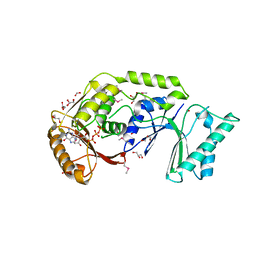 | | The crystal structure of a possible methylase from Clostridium difficile 630. | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tan, K, Wu, R, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a possible methylase from Clostridium difficile 630.
To be Published
|
|
3M1R
 
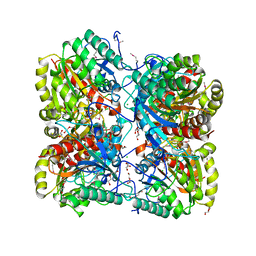 | | The crystal structure of formimidoylglutamase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Trevino, D, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-05 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The crystal structure of formimidoylglutamase from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
3MN2
 
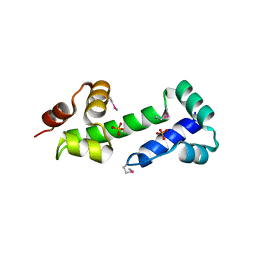 | |
3IKB
 
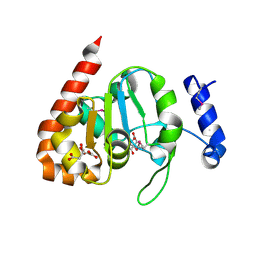 | |
3ILK
 
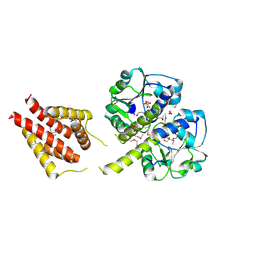 | | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20
To be Published
|
|
3OCJ
 
 | | The crystal structure of a possilbe exported protein from Bordetella parapertussis | | Descriptor: | GLYCEROL, PALMITIC ACID, Putative exported protein | | Authors: | Tan, K, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The crystal structure of a possilbe exported protein from Bordetella parapertussis
To be Published
|
|
3OM8
 
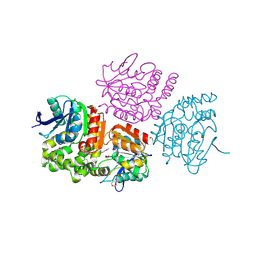 | | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable hydrolase | | Authors: | Tan, K, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01
To be Published
|
|
3ON3
 
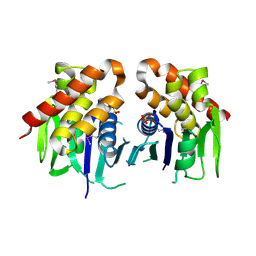 | | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA | | Descriptor: | Keto/oxoacid ferredoxin oxidoreductase, gamma subunit, SULFATE ION | | Authors: | Tan, K, Zhang, R, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA
To be Published
|
|
3OOS
 
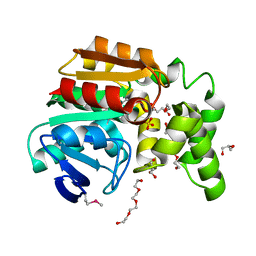 | | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne | | Descriptor: | Alpha/beta hydrolase family protein, GLYCEROL, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Bigelow, L, Hamilton, J, Li, H, Zhou, Y, Clancy, S, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne
To be Published
|
|
3N24
 
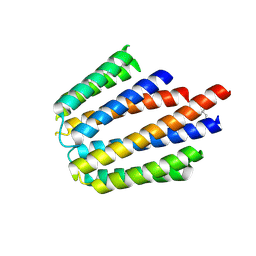 | |
3NZE
 
 | | The crystal structure of a domain of a possible sugar-binding transcriptional regulator from Arthrobacter aurescens TC1. | | Descriptor: | CALCIUM ION, Putative transcriptional regulator, sugar-binding family | | Authors: | Tan, K, Zhang, R, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The crystal structure of a domain of a possible sugar-binding transcriptional regulator from Arthrobacter aurescens TC1.
To be Published
|
|
3LL7
 
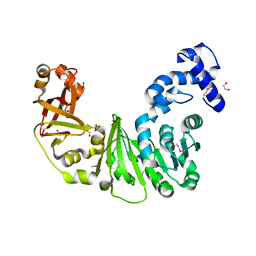 | | Crystal structure of putative methyltransferase PG_1098 from Porphyromonas gingivalis W83 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase | | Authors: | Chang, C, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-28 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative methyltransferase PG_1098 from Porphyromonas gingivalis W83
To be Published
|
|
3LEC
 
 | | The Crystal Structure of a protein in the NADB-Rossmann Superfamily from Streptococcus agalactiae to 1.8A | | Descriptor: | NADB-Rossmann Superfamily protein, SULFATE ION, ZINC ION | | Authors: | Stein, A.J, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of a protein in the NADB-Rossmann Superfamily from Streptococcus agalactiae to 1.8A
To be Published
|
|
3LR1
 
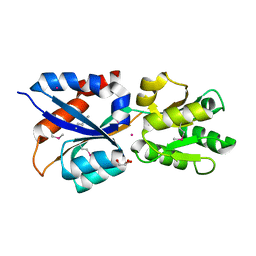 | | The crystal structure of the tungstate ABC transporter from Geobacter sulfurreducens | | Descriptor: | GLYCEROL, TUNGSTEN ION, Tungstate ABC transporter, ... | | Authors: | Zhang, R, Volkart, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the tungstate ABC transporter from Geobacter sulfurreducens
To be Published
|
|
3LAO
 
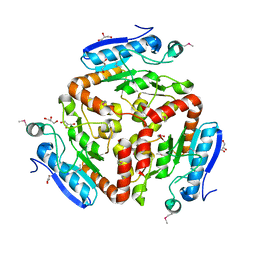 | |
3MFD
 
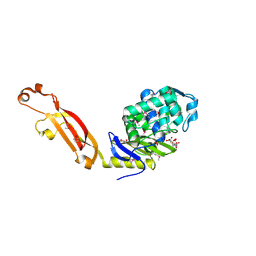 | | The Structure of the Beta-lactamase superfamily domain of D-alanyl-D-alanine carboxypeptidase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, D-alanyl-D-alanine carboxypeptidase dacB | | Authors: | Cuff, M.E, Rakowski, E, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-01 | | Release date: | 2010-05-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Beta-lactamase superfamily domain of D-alanyl-D-alanine carboxypeptidase from Bacillus subtilis.
TO BE PUBLISHED
|
|
3MQO
 
 | | The Crystal Structure of the PAS domain in complex with isopropanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.7A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Stein, A.J, Tesar, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the PAS domain in complex with isopropanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.7A
To be Published
|
|
3MQQ
 
 | | The Crystal Structure of the PAS domain in complex with Ethanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.65A | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, SODIUM ION, ... | | Authors: | Stein, A.J, Tesar, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the PAS domain in complex with Ethanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.65A
To be Published
|
|
