2GGF
 
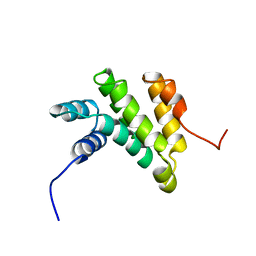 | | Solution structure of the MA3 domain of human Programmed cell death 4 | | Descriptor: | Programmed cell death 4, isoform 1 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-24 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MA3 domain of human Programmed cell death 4
To be Published
|
|
1IW7
 
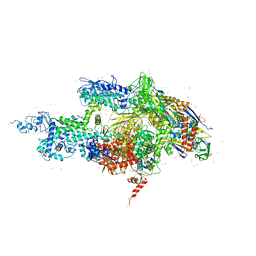 | |
1BKG
 
 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS WITH MALEATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, MALEIC ACID | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-07-07 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
1S0V
 
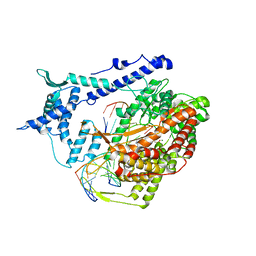 | | Structural basis for substrate selection by T7 RNA polymerase | | Descriptor: | 5'-D(*G*GP*GP*AP*AP*TP*CP*GP*AP*TP*AP*TP*CP*GP*CP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*AP*TP*TP*CP*CP*C)-3', 5'-R(*AP*AP*CP*U*GP*CP*GP*GP*CP*GP*AP*U)-3', ... | | Authors: | Temiakov, D, Patlan, V, Anikin, M, McAllister, W.T, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate selection by t7 RNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
2LEX
 
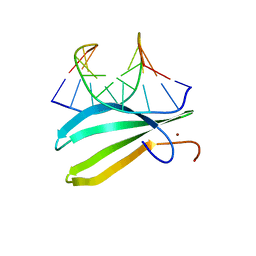 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | Descriptor: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | Authors: | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-24 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
1RRB
 
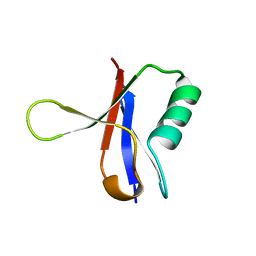 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
2BE5
 
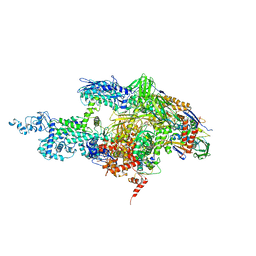 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with inhibitor tagetitoxin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Vassylyev, D.G, Svetlov, V, Vassylyeva, M.N, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Artsimovitch, I, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for transcription inhibition by tagetitoxin
Nat.Struct.Mol.Biol., 12, 2005
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
3JQJ
 
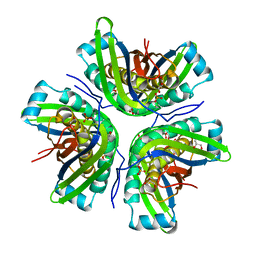 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | GLYCEROL, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KBB
 
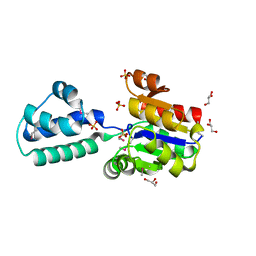 | | Crystal structure of putative beta-phosphoglucomutase from Thermotoga maritima | | Descriptor: | GLYCEROL, Phosphorylated carbohydrates phosphatase TM_1254, SULFATE ION | | Authors: | Strange, R.W, Antonyuk, S.V, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of a putative beta-phosphoglucomutase (TM1254) from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3KB6
 
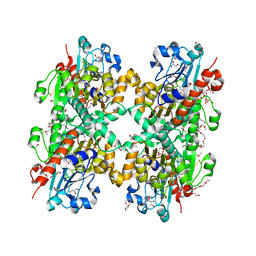 | | Crystal structure of D-Lactate dehydrogenase from aquifex aeolicus complexed with NAD and Lactic acid | | Descriptor: | D-lactate dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
8KCQ
 
 | | Solution structures of the N-terminal divergent caplonin homology (NN-CH) domains of human intraflagellar transport protein 54 | | Descriptor: | TRAF3-interacting protein 1 | | Authors: | Dang, W, Kuwasako, K, He, F, Takahashi, M, Tsuda, K, Nagata, T, Tanaka, A, Kobayashi, N, Kigawa, T, Guentert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2023-08-08 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structure of the N-terminal divergent calponin homology (NN-CH) domain of human intraflagellar transport protein 54.
Biomol.Nmr Assign., 18, 2024
|
|
7CKJ
 
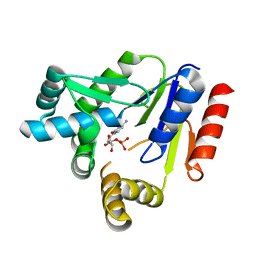 | | Crystal structure of CMP kinase in complex with CMP from Thermus thermophilus HB8 | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Cytidylate kinase | | Authors: | Mega, R, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of Thermus thermophilus CMP kinase complexed with a phosphoryl group acceptor and donor.
PLoS ONE, 15, 2020
|
|
3WND
 
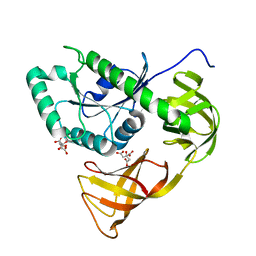 | | Crystal structure of EF-Pyl | | Descriptor: | CITRIC ACID, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
1D8K
 
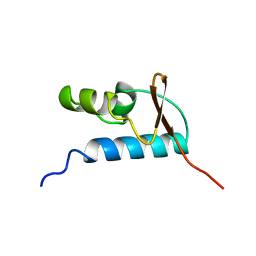 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1D8J
 
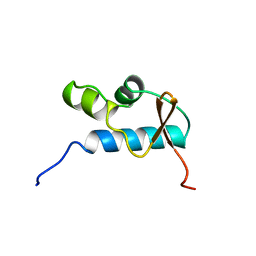 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1BJW
 
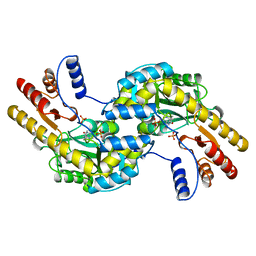 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-06-30 | | Release date: | 1999-07-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
1D2M
 
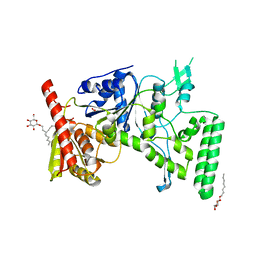 | | UVRB PROTEIN OF THERMUS THERMOPHILUS HB8; A NUCLEOTIDE EXCISION REPAIR ENZYME | | Descriptor: | EXCINUCLEASE ABC SUBUNIT B, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Nakagawa, N, Sugahara, M, Masui, R, Kato, R, Fukuyama, K, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-09-25 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair.
J.Biochem.(Tokyo), 126, 1999
|
|
1D9A
 
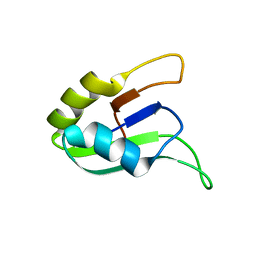 | | SOLUTION STRUCTURE OF THE SECOND RNA-BINDING DOMAIN (RBD2) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
1D8Z
 
 | | SOLUTION STRUCTURE OF THE FIRST RNA-BINDING DOMAIN (RBD1) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
5WVU
 
 | | Crystal structure of carboxypeptidase from Thermus thermophilus | | Descriptor: | GLYCEROL, Thermostable carboxypeptidase 1, ZINC ION | | Authors: | Okai, M, Nagata, K, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the transition between the open and closed conformations of Thermus thermophilus carboxypeptidase.
Biochem. Biophys. Res. Commun., 484, 2017
|
|
1J1V
 
 | | Crystal structure of DnaA domainIV complexed with DnaAbox DNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*TP*GP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*TP*AP*TP*CP*CP*AP*CP*AP*GP*G)-3', Chromosomal replication initiator protein dnaA | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Terada, T, Shirouzu, M, Katayama, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-18 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of replication origin recognition by the DnaA protein
NUCLEIC ACIDS RES., 31, 2003
|
|
7FBR
 
 | |
