7SJY
 
 | |
4LVW
 
 | |
2XEU
 
 | | Ring domain | | Descriptor: | RING FINGER PROTEIN 4, SULFATE ION, ZINC ION, ... | | Authors: | Plechanovova, A, McMahon, S.A, Johnson, K.A, Navratilova, I, Naismith, J.H, Hay, R.T. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of Ubiquitylation by Dimeric Ring Ligase Rnf4
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XO0
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to 24-diamino-1,3,5- triazine identified by virtual screening | | Descriptor: | 1,3,5-TRIAZINE-2,4-DIAMINE, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
3K0X
 
 | | Crystal structure of telomere capping protein Ten1 from Saccharomyces pombe | | Descriptor: | IODIDE ION, Protein Ten1 | | Authors: | Gelinas, A.D, Reyes, F.E, Batey, R.T, Wuttke, D.S. | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4LE4
 
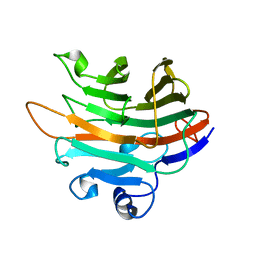 | | Crystal structure of PaGluc131A with cellotriose | | Descriptor: | Beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, T, Chan, H.C, Huang, C.H, Ko, T.P, Huang, T.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a GH131 beta-Glucanase Catalytic Domain from Podospora anserina in Complex with Cellotriose
To be Published
|
|
4LE3
 
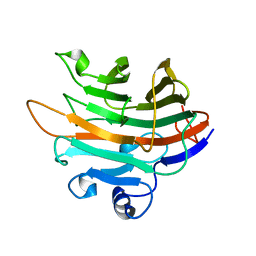 | | Crystal structure of a GH131 beta-glucanase catalytic domain from Podospora anserina | | Descriptor: | Beta-glucanase | | Authors: | Jiang, T, Chan, H.C, Huang, C.H, Ko, T.P, Huang, T.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a GH131 beta-Glucanase Catalytic Domain from Podospora anserina in Complex with Cellotriose
To be Published
|
|
3KB7
 
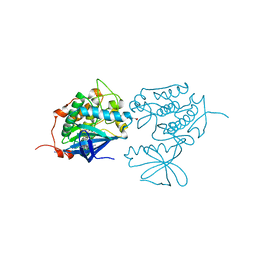 | | Crystal structure of Polo-like kinase 1 in complex with a pyrazoloquinazoline inhibitor | | Descriptor: | 8-{[2-methoxy-5-(4-methylpiperazin-1-yl)phenyl]amino}-1-methyl-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxamide, L(+)-TARTARIC ACID, Serine/threonine-protein kinase PLK1, ... | | Authors: | Bossi, R.T, Bertrand, J.A. | | Deposit date: | 2009-10-20 | | Release date: | 2010-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of 4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline derivatives as a new class of orally and selective Polo-like kinase 1 inhibitors
J.Med.Chem., 53, 2010
|
|
3RSP
 
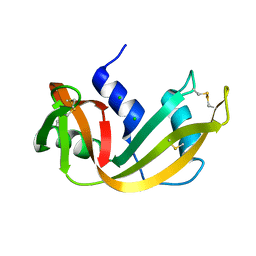 | | STRUCTURE OF THE P93G VARIANT OF RIBONUCLEASE A | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A | | Authors: | Schultz, L.W, Hargraves, S.R, Klink, T.A, Raines, R.T. | | Deposit date: | 1997-10-20 | | Release date: | 1998-04-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability of the P93G variant of ribonuclease A.
Protein Sci., 7, 1998
|
|
2KW8
 
 | |
2LBZ
 
 | | Thurincin H | | Descriptor: | Thuricin17 | | Authors: | Sit, C.S, van Belkum, M.J, Mckay, R.T, Worobo, R.W, Vederas, J.C. | | Deposit date: | 2011-04-10 | | Release date: | 2012-01-18 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D solution structure of thurincin H, a bacteriocin with four sulfur to alpha-carbon crosslinks.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2LN7
 
 | |
2LA0
 
 | | Trn- peptide of the two-component bacteriocin Thuricin CD | | Descriptor: | Uncharacterized protein | | Authors: | Sit, C.S, Mckay, R.T, Hill, C, Ross, R.P, Vederas, J.C. | | Deposit date: | 2011-02-27 | | Release date: | 2012-01-11 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links.
J.Am.Chem.Soc., 133, 2011
|
|
7JR6
 
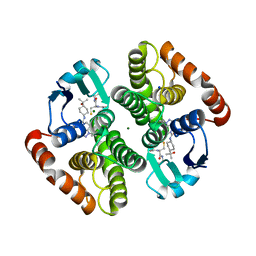 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1-(3-fluorophenyl)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7JR8
 
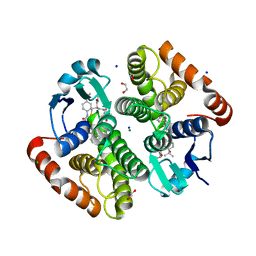 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
830C
 
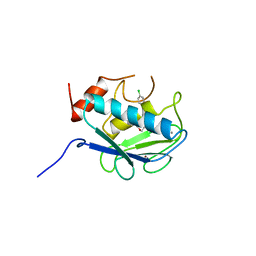 | | COLLAGENASE-3 (MMP-13) COMPLEXED TO A SULPHONE-BASED HYDROXAMIC ACID | | Descriptor: | 4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYLMETHYL]-TETRAHYDRO-PYRAN-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
6P2H
 
 | |
6IJ1
 
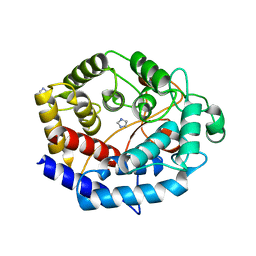 | | Crystal structure of a protein from Actinoplanes | | Descriptor: | ACETATE ION, IMIDAZOLE, Prenylcyclase | | Authors: | Yang, Z.Z, Zhang, L.L, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-10-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Crystal structure of TchmY from Actinoplanes teichomyceticus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6PP7
 
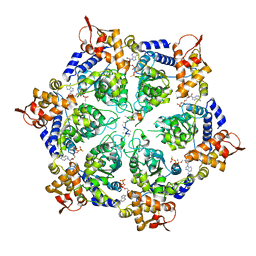 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6ITA
 
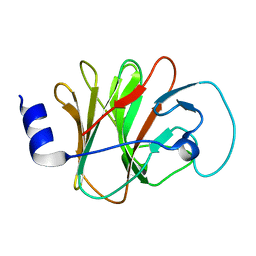 | | Crystal structure of intracellular B30.2 domain of BTN3A1 mutant | | Descriptor: | Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-11-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
7QPB
 
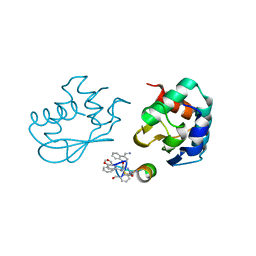 | | Catalytic C-lobe of the HECT-type ubiquitin ligase E6AP in complex with a hybrid foldamer-peptide macrocycle | | Descriptor: | Isoform I of Ubiquitin-protein ligase E3A, hybrid foldamer-peptide macrocycle | | Authors: | Dengler, S, Howard, R.T, Morozov, V, Tsiamantas, C, Douat, C, Suga, H, Huc, I. | | Deposit date: | 2022-01-03 | | Release date: | 2023-09-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Display Selection of a Hybrid Foldamer-Peptide Macrocycle.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5XFY
 
 | | Crystal structure of a novel PET hydrolase S131A mutant from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XG0
 
 | | Crystal structure of a novel PET hydrolase from Ideonella sakaiensis 201-F6 | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XK6
 
 | | Structure of a prenyltransferase soaked with IPP | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, SULFATE ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XFZ
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
