1M5Y
 
 | |
3BVO
 
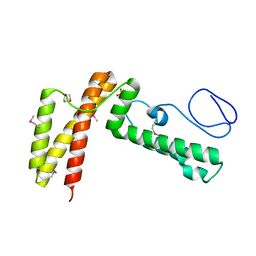 | | Crystal structure of human co-chaperone protein HscB | | Descriptor: | Co-chaperone protein HscB, mitochondrial precursor, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-01-07 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain.
J.Biol.Chem., 283, 2008
|
|
4KX3
 
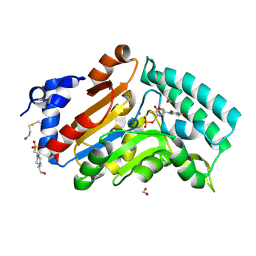 | |
4KX5
 
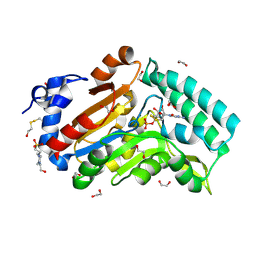 | |
1U4G
 
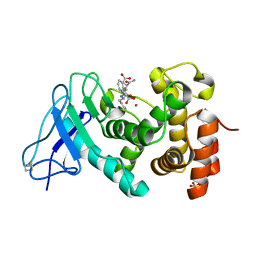 | | Elastase of Pseudomonas aeruginosa with an inhibitor | | Descriptor: | CALCIUM ION, Elastase, N-(1-CARBOXY-3-PHENYLPROPYL)PHENYLALANYL-ALPHA-ASPARAGINE, ... | | Authors: | Bitto, E, McKay, D.B. | | Deposit date: | 2004-07-25 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Elastase of Pseudomonas aeruginosa with an inhibitor
To be Published, 2004
|
|
2QNK
 
 | | Crystal structure of human 3-hydroxyanthranilate 3,4-dioxygenase | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, NICKEL (II) ION, PHOSPHATE ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-07-18 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Human 3-hydroxyanthranilate 3,4-dioxygenase.
to be published
|
|
3C9Q
 
 | | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide.
To be Published
|
|
2GU2
 
 | | Crystal Structure of an Aspartoacylase from Rattus norvegicus | | Descriptor: | Aspa protein, SULFATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-06-20 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2IFU
 
 | | Crystal Structure of a Gamma-SNAP from Danio rerio | | Descriptor: | SULFATE ION, gamma-snap | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family.
Proteins, 70, 2008
|
|
2NXF
 
 | | Crystal Structure of a dimetal phosphatase from Danio rerio LOC 393393 | | Descriptor: | ETHANOL, PHOSPHATE ION, Putative dimetal phosphatase, ... | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a dimetal phosphatase from Danio rerio LOC 393393
To be Published
|
|
2OA1
 
 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the L-Tryptophan with FAD complex | | Descriptor: | ADENOSINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bitto, E, Bingman, C.A, Singh, S, Phillips Jr, G.N. | | Deposit date: | 2006-12-14 | | Release date: | 2007-04-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
2G09
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product complex | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2I3C
 
 | | Crystal Structure of an Aspartoacylase from Homo Sapiens | | Descriptor: | Aspartoacylase, PHOSPHATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2NYI
 
 | | Crystal Structure of an Unknown Protein from Galdieria sulphuraria | | Descriptor: | unknown protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tandem ACT domain-containing protein ACTP from Galdieria sulphuraria.
Proteins, 80, 2012
|
|
2G0A
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1 with lead(II) bound in active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III, LEAD (II) ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2O9Z
 
 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the Apo form | | Descriptor: | PHOSPHATE ION, Tryptophan halogenase | | Authors: | Bitto, E, Bingman, C.A, Phillips Jr, G.N. | | Deposit date: | 2006-12-14 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
2G06
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, with bound magnesium(II) | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2G07
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, phospho-enzyme intermediate analog with Beryllium fluoride | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2H1S
 
 | |
2G08
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product-transition complex analog with Aluminum fluoride | | Descriptor: | ALUMINUM FLUORIDE, Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
4W79
 
 | | Crystal Structure of Human Protein N-terminal Glutamine Amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Protein N-terminal glutamine amidohydrolase, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human protein N-terminal glutamine amidohydrolase, an initial component of the N-end rule pathway.
Plos One, 9, 2014
|
|
4FE3
 
 | |
2BE4
 
 | | X-RAY STRUCTURE AN EF-HAND PROTEIN FROM DANIO RERIO Dr.36843 | | Descriptor: | hypothetical protein LOC449832 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-21 | | Release date: | 2005-11-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of Danio rerio secretagogin: A hexa-EF-hand calcium sensor.
Proteins, 76, 2009
|
|
2BDU
 
 | | X-Ray Structure of a Cytosolic 5'-Nucleotidase III from Mus Musculus MM.158936 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
3D0R
 
 | | Crystal structure of calG3 from Micromonospora echinospora determined in space group P2(1) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Protein CalG3 | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N. | | Deposit date: | 2008-05-02 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
Chem.Biol., 15, 2008
|
|
