1NEZ
 
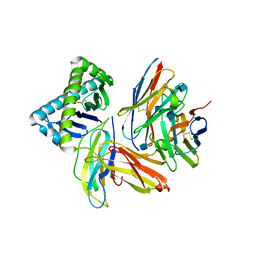 | | The Crystal Structure of a TL/CD8aa Complex at 2.1A resolution:Implications for Memory T cell Generation, Co-receptor Preference and Affinity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, Y, Xiong, Y, Naidenko, O.V, Liu, J.H, Zhang, R, Joachimiak, A, Kronenberg, M, Cheroutre, H, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2002-12-12 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a TL/CD8alphaalpha Complex at 2.1 A resolution: Implications for modulation of T cell activation and memory
Immunity, 18, 2003
|
|
4OR4
 
 | |
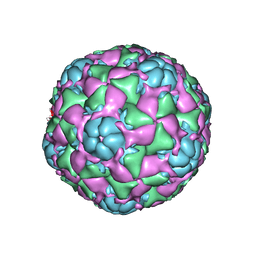
6CRS
 
 | |
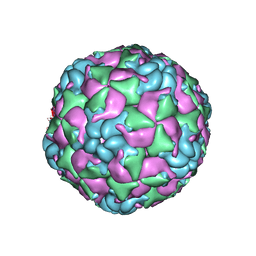
6CRU
 
 | |
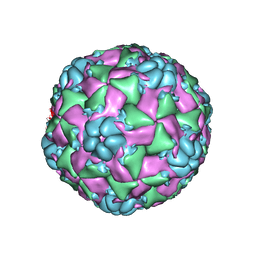
6CS4
 
 | |
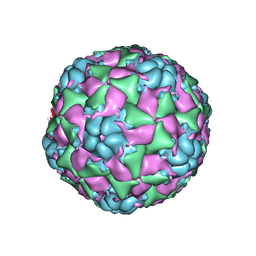
6CS6
 
 | |
4WM7
 
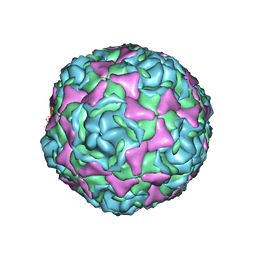 | | Crystal Structure of Human Enterovirus D68 in Complex with Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, VP1, VP2, ... | | Authors: | Liu, Y, Sheng, J, Fokine, A, Meng, G, Long, F, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2014-10-08 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Virus structure. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children.
Science, 347, 2015
|
|
3VAC
 
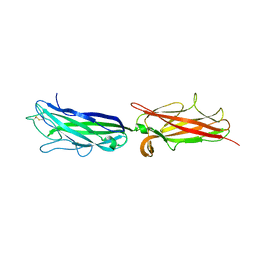 | |
8AHL
 
 | |
8AFH
 
 | |
8AJB
 
 | |
8AIA
 
 | |
8AFE
 
 | |
8AIX
 
 | |
8AFL
 
 | |
8AFM
 
 | |
6X9H
 
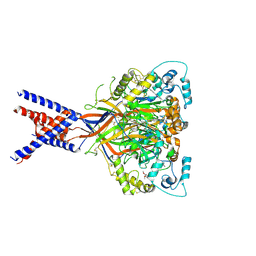 | | Molecular mechanism and structural basis of small-molecule modulation of acid-sensing ion channel 1 (ASIC1) | | Descriptor: | 2-[4-(3,4-dimethoxyphenoxy)phenyl]-1H-benzimidazole-6-carboximidamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, ... | | Authors: | Liu, Y, Ma, J, DesJarlais, R.L, Hagan, R, Rech, J, Lin, D, Liu, C, Miller, R, Schoellerman, J, Luo, J, Letavic, M, Grasberger, B, Maher, M. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular mechanism and structural basis of small-molecule modulation of the gating of acid-sensing ion channel 1.
Commun Biol, 4, 2021
|
|
1JS0
 
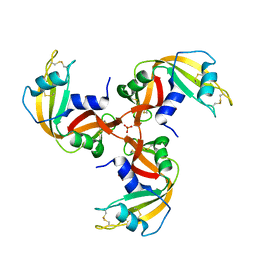 | | Crystal Structure of 3D Domain-swapped RNase A Minor Trimer | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Liu, Y, Gotte, G, Libonati, M, Eisenberg, D. | | Deposit date: | 2001-08-15 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the two 3D domain-swapped RNase A trimers.
Protein Sci., 11, 2002
|
|
3GYO
 
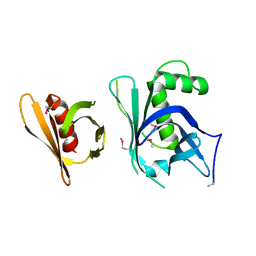 | | Se-Met Rtt106p | | Descriptor: | Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
3GYP
 
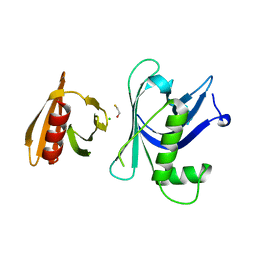 | | Rtt106p | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
1RIM
 
 | | E6-binding zinc finger (E6apc2) | | Descriptor: | E6apc2 peptide | | Authors: | Liu, Y, Liu, Z, Androphy, E, Chen, J, Baleja, J.D. | | Deposit date: | 2003-11-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of helical peptides that inhibit the E6 protein of papillomavirus.
Biochemistry, 43, 2004
|
|
1RIJ
 
 | | E6-bind Trp-cage (E6apn1) | | Descriptor: | E6apn1 peptide | | Authors: | Liu, Y, Liu, Z, Androphy, E, Chen, J, Baleja, J.D. | | Deposit date: | 2003-11-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of helical peptides that inhibit the E6 protein of papillomavirus.
Biochemistry, 43, 2004
|
|
1RIK
 
 | | E6-binding zinc finger (E6apc1) | | Descriptor: | E6apc1 peptide | | Authors: | Liu, Y, Liu, Z, Androphy, E, Chen, J, Baleja, J.D. | | Deposit date: | 2003-11-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of helical peptides that inhibit the E6 protein of papillomavirus.
Biochemistry, 43, 2004
|
|
1C8J
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM MUTANT (F87W/Y96F) | | Descriptor: | CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, Y, Jiang, F, Guo, Q, Chen, X, Jin, J, Sun, Y, Rao, Z. | | Deposit date: | 2000-05-31 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytochrome P450cam mutant (F87W/Y96F)
To be Published
|
|
3OIX
 
 | | Crystal structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Putative dihydroorotate dehydrogenase; dihydroorotate oxidase | | Authors: | Liu, Y, Gao, Z.Q, Liu, C.P, Dong, Y.H. | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans
Acta Crystallogr.,Sect.F, 67, 2011
|
|
