1R7Z
 
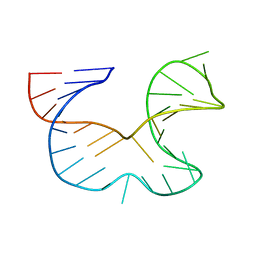 | | NMR STRUCTURE OF THE R(GGAGGACAUUCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUUCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
4PO3
 
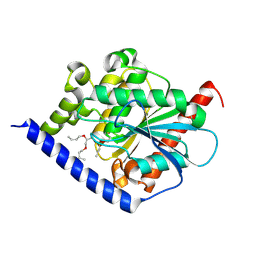 | |
3ZXZ
 
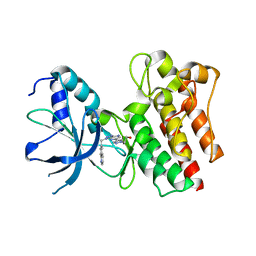 | | X-ray Structure of PF-04217903 bound to the kinase domain of c-Met | | Descriptor: | 2-{4-[1-(QUINOLIN-6-YLMETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRAZIN-6-YL]-1H-PYRAZOL-1-YL}ETHANOL, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-16 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
4L8M
 
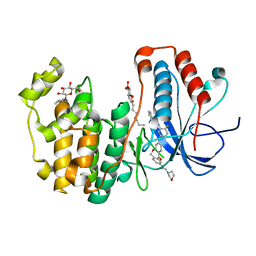 | | Human p38 MAP kinase in complex with a Dibenzoxepinone | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-fluoro-5-({9-[2-(morpholin-4-yl)ethoxy]-11-oxo-6,11-dihydrodibenzo[b,e]oxepin-3-yl}amino)phenyl]benzamide, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2013-06-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metabolically Stable Dibenzo[b,e]oxepin-11(6H)-ones as Highly Selective p38 MAP Kinase Inhibitors: Optimizing Anti-Cytokine Activity in Human Whole Blood.
J.Med.Chem., 56, 2013
|
|
3SDL
 
 | | Crystal structure of human ISG15 in complex with NS1 N-terminal region from influenza B virus, Northeast Structural Genomics Consortium Target IDs HX6481, HR2873, and OR2 | | Descriptor: | Non-structural protein 1, Ubiquitin-like protein ISG15 | | Authors: | Guan, R, Ma, L.-C, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza B virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
216D
 
 | |
217D
 
 | |
4RIO
 
 | |
4B0G
 
 | | Complex of Aurora-A bound to an Imidazopyridine-based inhibitor | | Descriptor: | 6-bromo-2-(1-methyl-1H-imidazol-5-yl)-7-{4-[(5-methyl-1,2-oxazol-3-yl)methyl]piperazin-1-yl}-1H-imidazo[4,5-b]pyridine, AURORA KINASE A, SULFATE ION | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2012-07-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of Imidazo[4,5-B]Pyridine-Based Kinase Inhibitors: Identification of a Dual Flt3/Aurora Kinase Inhibitor as an Orally Bioavailable Preclinical Development Candidate for the Treatment of Acute Myeloid Leukemia.
J.Med.Chem., 55, 2012
|
|
4HFX
 
 | | Crystal structure of a transcription elongation factor B polypeptide 3 from Homo sapiens, Northeast Structural Genomics consortium target id HR4748B. | | Descriptor: | SULFATE ION, Transcription elongation factor B polypeptide 3 | | Authors: | Seetharaman, J, Su, M, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-05 | | Release date: | 2012-12-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a transcription elongation factor B polypeptide 3 from Homo sapiens, Northeast Structural Genomics consortium target id HR4748B. (CASP Target)
TO BE PUBLISHED
|
|
3FER
 
 | | Crystal structure of n-terminal actin-binding domain from human filamin b (tandem ch-domains). northeast structural genomics consortium target hr5571a. | | Descriptor: | ACETIC ACID, Filamin-B | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, R, Shastry, R, Sahdev, S, Ciccosanti, C, Xiao, R, Everett, J.K, Huang, Y, Acton, T, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-30 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of n-terminal actin-binding domain from human filamin b (tandem ch-domains). northeast structural genomics consortium target hr5571a.
To be Published
|
|
4PY1
 
 | | Crystal structure of Tyk2 in complex with compound 15, 6-((2,5-dimethoxyphenyl)thio)-3-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine | | Descriptor: | 6-[(2,5-dimethoxyphenyl)sulfanyl]-3-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazine, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Kinase domain inhibition of leucine rich repeat kinase 2 (LRRK2) using a [1,2,4]triazolo[4,3-b]pyridazine scaffold.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2GPB
 
 | |
1QCX
 
 | | PECTIN LYASE B | | Descriptor: | PECTIN LYASE B | | Authors: | Vitali, J, Jurnak, F. | | Deposit date: | 1999-05-13 | | Release date: | 1999-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The tree-dimensional structure of aspergillus niger pectin lyase B at 1.7-A resolution.
Plant Physiol., 116, 1998
|
|
1RW8
 
 | | Crystal Structure of TGF-beta receptor I kinase with ATP site inhibitor | | Descriptor: | 3-(4-FLUOROPHENYL)-2-(6-METHYLPYRIDIN-2-YL)-5,6-DIHYDRO-4H-PYRROLO[1,2-B]PYRAZOLE, TGF-beta receptor type I | | Authors: | Zhang, F, Sawyer, J.S. | | Deposit date: | 2003-12-16 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
3LQK
 
 | | Crystal structure of dipicolinate synthase subunit B from Bacillus halodurans C | | Descriptor: | Dipicolinate synthase subunit B, PHOSPHATE ION | | Authors: | Nocek, B, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-09 | | Release date: | 2010-03-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dipicolinate synthase subunit B from Bacillus halodurans C
To be Published
|
|
4QL8
 
 | | Crystal structure of Androgen Receptor in complex with the ligand | | Descriptor: | 2-chloro-4-[(3S,3aS,4S)-4-hydroxy-3-methoxy-3a,4,5,6-tetrahydro-3H-pyrrolo[1,2-b]pyrazol-2-yl]-3-methylbenzonitrile, Androgen receptor | | Authors: | Krishnamurthy, N, Sangeetha, R, Ghadiyaram, C, Sasmal, S, Subramanya, H.S. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3-alkoxy-pyrrolo[1,2-b]pyrazolines as selective androgen receptor modulators with ideal physicochemical properties for transdermal administration
J.Med.Chem., 57, 2014
|
|
3GPJ
 
 | | Crystal structure of the yeast 20S proteasome in complex with syringolin B | | Descriptor: | N-{[(1S)-2-methyl-1-{[(5S,8S)-5-(1-methylethyl)-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]carbamoyl}propyl]carbamoyl}-L-valine, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Huber, R, Kaiser, M. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic and structural studies on syringolin A and B reveal critical determinants of selectivity and potency of proteasome inhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5E
 
 | | Human aldose reductase complexed with IDD 740 inhibitor | | Descriptor: | 2-(3-((4,5,7-trifluorobenzo[d]thiazol-2-yl)methyl)-1H-pyrrolo[2,3-b]pyridin-1-yl)acetic acid, Aldose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Podjarny, A.D, Van Zandt, M.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of [3-(4,5,7-trifluoro-benzothiazol-2-ylmethyl)-pyrrolo[2,3-b]pyridin-1-yl]acetic acids as highly potent and selective inhibitors of aldose reductase for treatment of chronic diabetic complications.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4PB9
 
 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab64 | | Descriptor: | Ab64 heavy chain, Ab64 light chain, SULFATE ION | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-11 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
4PB0
 
 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab53 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ab53 heavy chain, Ab53 light chain, ... | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
5N1X
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-ethyl-5-pyridin-3-yl-pyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1Z
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine macrocyclic ligand | | Descriptor: | B-cell lymphoma 6 protein, CHLORIDE ION, pyrazolo-pyrimidine macrocycle | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
6EW7
 
 | |
5N20
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-2-[(3~{R})-3-(dimethylamino)pyrrolidin-1-yl]phenyl]ethanamide | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
