4FQK
 
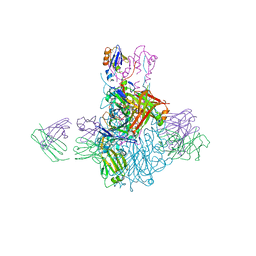 | | Influenza B/Brisbane/60/2008 hemagglutinin Fab CR8059 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody CR8059 Heavy Chain, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (5.65 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQJ
 
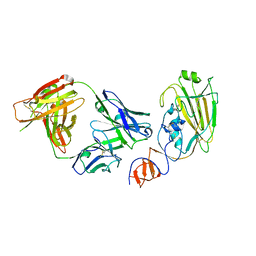 | | Influenza B/Florida/4/2006 hemagglutinin Fab CR8071 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, antibody CR8071 heavy chain, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4GKO
 
 | | Crystal structure of the calcium2+-bound human IgE-Fc(epsilon)3-4 bound to its B cell receptor derCD23 | | Descriptor: | CALCIUM ION, Ig epsilon chain C region, Low affinity immunoglobulin epsilon Fc receptor, ... | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
6BVF
 
 | |
6BVN
 
 | |
1K06
 
 | | Crystallographic Binding Study of 100 mM N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b | | Descriptor: | Glycogen Phosphorylase, N-[(phenylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2001-09-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of N-acetyl-N '-beta-D-glucopyranosyl urea and N-benzoyl-N '-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
1JO2
 
 | |
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
1K08
 
 | | Crystallographic Binding Study of 10 mM N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b | | Descriptor: | Glycogen Phosphorylase, N-[(phenylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2001-09-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Binding of N-acetyl-N '-beta-D-glucopyranosyl urea and N-benzoyl-N '-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
3OPI
 
 | | 7-DEAZA-2'-DEOXYADENOSINE modification in B-FORM DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*(7DA)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, SODIUM ION | | Authors: | Kowal, E.A, Ganguly, M, Pallan, P.S, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | Deposit date: | 2010-09-01 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Altering the Electrostatic Potential in the Major Groove: Thermodynamic and Structural Characterization of 7-Deaza-2'-deoxyadenosine:dT Base Pairing in DNA.
J.Phys.Chem.B, 115, 2011
|
|
3JV6
 
 | |
5ACC
 
 | | A Novel Oral Selective Estrogen Receptor Down-regulator, AZD9496, drives Tumour Growth Inhibition in Estrogen Receptor positive and ESR1 Mutant Models | | Descriptor: | (E)-3-(3,5-DIFLUORO-4-((1R,3R)-2-(2-FLUORO-2- METHYLPROPYL)-3-METHYL-2,3,4,9-TETRAHYDRO-1H-PYRIDO(3,4-B)INDOL-1-YL)PHENYL)ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Weir, H.M, Bradbury, R.H, Lawson, M, Rabow, A.A, Buttar, D, Callis, R.J, Curwen, J.O, de Almeida, C, Ballard, P, Hulse, M, Donald, C.S, Feron, L.J.L, Gingell, H, Karoutchi, G, MacFaul, P, Moss, T, Pearson, S.E, Tonge, M, Davies, G, Walker, G.E, Wilson, Z, Rowlinson, R, Powell, S, Hemsley, P, Linney, E, Campbell, H, Ghazoui, Z, Sadler, C, Richmond, G, Pazolli, E, Mazzola, A.M, DCruz, C, De Savi, C. | | Deposit date: | 2015-08-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
4IWD
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-8033 analog | | Descriptor: | 1-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)methanesulfonamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Northrup, A, Rickert, K, Patel, S, Allison, T. | | Deposit date: | 2013-01-23 | | Release date: | 2013-12-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met.
J.Med.Chem., 56, 2013
|
|
4J6K
 
 | |
1XHA
 
 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-{4-[(4-{3-[(2R)-3,3-DIMETHYLPIPERIDIN-2-YL]-2-FLUORO-6-HYDROXYBENZOYL}BENZOYL)AMINO]AZEPAN-3-YL}ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1XH8
 
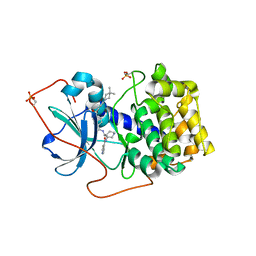 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-[4-({4-[5-(4,4-DIMETHYLPIPERIDIN-1-YL)-2-HYDROXYBENZOYL]BENZOYL}AMINO)AZEPAN-3-YL]ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1XH6
 
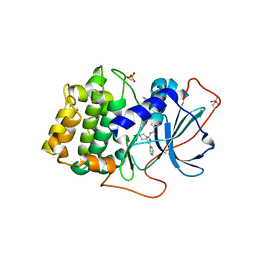 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-(4-{[4-(2-HYDROXY-5-PIPERIDIN-1-YLBENZOYL)BENZOYL]AMINO}AZEPAN-3-YL)ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1XH7
 
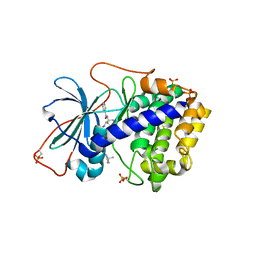 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-[4-({4-[5-(3,3-DIMETHYLPIPERIDIN-1-YL)-2-HYDROXYBENZOYL]BENZOYL}AMINO)AZEPAN-3-YL]ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1XH5
 
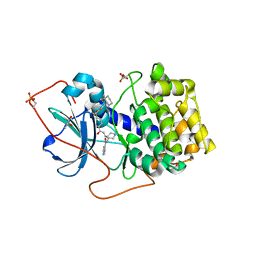 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-{4-[(4-{3-[(2R)-3,3-DIMETHYLPIPERIDIN-2-YL]-2-FLUORO-6-HYDROXYBENZOYL}BENZOYL)AMINO]AZEPAN-3-YL}ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1XH9
 
 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-[4-({4-[5-(DIMETHYLAMINO)-2-HYDROXYBENZOYL]BENZOYL}AMINO)AZEPAN-3-YL]ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1XH4
 
 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-[4-({4-[5-(DIMETHYLAMINO)-2-HYDROXYBENZOYL]BENZOYL}AMINO)AZEPAN-3-YL]ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
2R76
 
 | | Crystal structure of the rare lipoprotein B (SO_1173) from Shewanella oneidensis, Northeast Structural Genomics Consortium Target SoR91A | | Descriptor: | Rare lipoprotein B | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Maglaqui, M, Owen, L.A, Cunningham, K, Fang, Y, Xiao, R, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the rare lipoprotein B (SO_1173) from Shewanella oneidensis.
To be Published
|
|
1YHT
 
 | | Crystal structure analysis of Dispersin B | | Descriptor: | ACETIC ACID, DspB, GLYCEROL | | Authors: | Ramasubbu, N, Thomas, L.M, Ragunath, C, Kaplan, J.B. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Dispersin B, a Biofilm-releasing Glycoside Hydrolase from the Periodontopathogen Actinobacillus actinomycetemcomitans.
J.Mol.Biol., 349, 2005
|
|
3RRL
 
 | | Complex structure of 3-oxoadipate coA-transferase subunit A and B from Helicobacter pylori 26695 | | Descriptor: | GLYCEROL, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit A, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit B | | Authors: | Nocek, B, Stein, A, Marshall, N, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-06-29 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Complex structure of 3-oxoadipate coA-transferase subunit A and B
from Helicobacter pylori 26695
TO BE PUBLISHED
|
|
4FN5
 
 | |
