3NW4
 
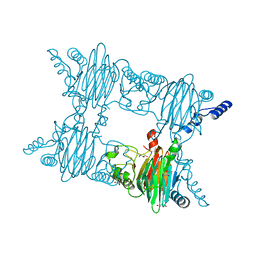 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans in complex with gentisate | | Descriptor: | 2,5-dihydroxybenzoic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
5U55
 
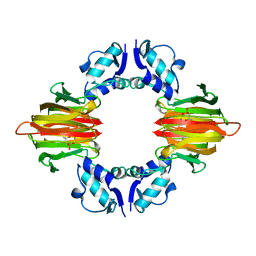 | | Psf4 in complex with Mn2+ and (S)-2-HPP | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, (S)-2-hydroxypropylphosphonic acid epoxidase, MANGANESE (II) ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
5U5D
 
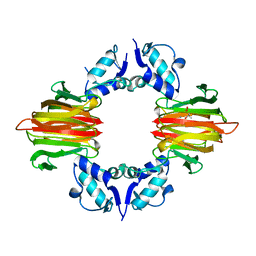 | | Psf4 in complex with Mn2+ and (R)-2-HPP | | Descriptor: | (S)-2-hydroxypropylphosphonic acid epoxidase, MANGANESE (II) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
5TG0
 
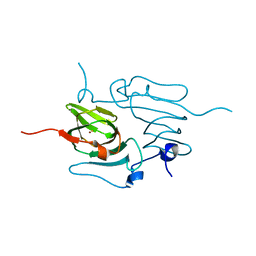 | |
5U58
 
 | | Psf4 in complex with Fe2+ and (R)-2-HPP | | Descriptor: | (S)-2-hydroxypropylphosphonic acid epoxidase, FE (III) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
3NJZ
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans Adducts with salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-06-18 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: A step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases.
J.Struct.Biol., 177, 2012
|
|
3NL1
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans Adducts with gentisate | | Descriptor: | 2,5-dihydroxybenzoic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: A step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases.
J.Struct.Biol., 177, 2012
|
|
5UQP
 
 | | The crystal structure of cupin protein from Rhodococcus jostii RHA1 | | Descriptor: | CHLORIDE ION, Cupin, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cupin protein from Rhodococcus jostii RHA1
To Be Published
|
|
2DCT
 
 | | Crystal structure of the TT1209 from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, SODIUM ION, hypothetical protein TTHA0104 | | Authors: | Asada, Y, Sugahara, M, Shimizu, K, Yamamoto, H, Shimada, H, Nakamoto, T, Ono, N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-12 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the TT1209 from Thermus thermophilus HB8
To be Published
|
|
2D40
 
 | |
2F4P
 
 | |
3H7J
 
 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in monoclinic form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Role of Bacillus subtilis BacB in the synthesis of bacilysin
J.Biol.Chem., 284, 2009
|
|
3H50
 
 | |
3H7Y
 
 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in tetragonal form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Role of Bacillus subtilis BacB in the synthesis of bacilysin
J.Biol.Chem., 284, 2009
|
|
3I7D
 
 | |
3IBM
 
 | | CRYSTAL STRUCTURE OF cupin 2 domain-containing protein Hhal_0468 FROM Halorhodospira halophila | | Descriptor: | Cupin 2, conserved barrel domain protein, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-16 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF cupin 2 domain-containing PROTEIN Hhal_0468 FROM Halorhodospira halophila
To be Published
|
|
3JZV
 
 | |
3HT1
 
 | |
3HT2
 
 | |
3H9A
 
 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in triclinic form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-30 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Analysis of multiple crystal forms of Bacillus subtilis BacB suggests a role for a metal ion as a nucleant for crystallization
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3H8U
 
 | |
3KGZ
 
 | | Crystal structure of a cupin 2 conserved barrel domain protein from Rhodopseudomonas palustris | | Descriptor: | Cupin 2 conserved barrel domain protein, MANGANESE (II) ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-29 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a cupin 2 conserved barrel domain protein from Rhodopseudomonas palustris
To be Published
|
|
8E8W
 
 | |
6XCV
 
 | |
6VZY
 
 | |
