2YBD
 
 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 with bound phosphate | | Descriptor: | HYDROLASE, HALOACID DEHALOGENASE-LIKE FAMILY, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable Had Family Hydrolase from Pseudomonas Fluorescens Pf-5 with Bound Phosphate
To be Published
|
|
2IM9
 
 | | Crystal structure of protein LPG0564 from Legionella pneumophila str. Philadelphia 1, Pfam DUF1460 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of hypothetical protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
TO BE PUBLISHED
|
|
2KUC
 
 | | Solution Structure of a putative disulphide-isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Putative disulphide-isomerase | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-02 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a putative disulphide-isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
2L5L
 
 | | Solution Structure of Thioredoxin from Bacteroides Vulgatus | | Descriptor: | Thioredoxin | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-02 | | Release date: | 2010-11-24 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Thioredoxin from Bacteroides Vulgatus
To be Published
|
|
2L5O
 
 | | Solution Structure of a Putative Thioredoxin from Neisseria meningitidis | | Descriptor: | Putative thioredoxin | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-03 | | Release date: | 2010-12-15 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Putative Thioredoxin from Neisseria meningitidis
To be Published
|
|
2L57
 
 | | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens
To be Published
|
|
2IA1
 
 | | Crystal structure of protein BH3703 from Bacillus halodurans, Pfam DUF600 | | Descriptor: | BH3703 protein, GLYCEROL, SULFATE ION | | Authors: | Ramagopal, U.A, Russell, M, Toro, R, Freeman, J.C, Reyes, C, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of hypothetical protein BH3703 from Bacillus halodurans
To be Published
|
|
3N05
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM STREPTOMYCES AVERMITILIS | | Descriptor: | NH(3)-dependent NAD(+) synthetase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Nh3-Dependent Nad+ Synthetase from Streptomyces Avermitilis
To be Published
|
|
3L60
 
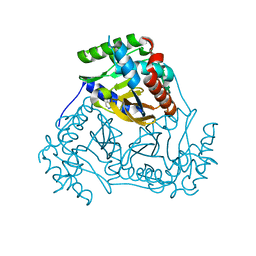 | | Crystal structure of branched-chain alpha-keto acid dehydrogenase subunit e2 from mycobacterium tuberculosis | | Descriptor: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE, UNKNOWN LIGAND | | Authors: | Zencheck, W.D, Bonanno, J.B, Patskovsky, Y, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Branched-Chain Alpha-Keto Acid Dehydrogenase Subunit E2 from Mycobacterium Tuberculosis
To be Published
|
|
3NRJ
 
 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a complexed with magnesium | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Had Family Hydrolase from Pseudomonas Syringae Pv.Phaseolica 1448A
To be Published
|
|
3R09
 
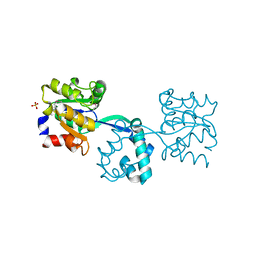 | | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg | | Descriptor: | Hydrolase, haloacid dehalogenase-like family, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-03-07 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg
To be Published
|
|
3MGK
 
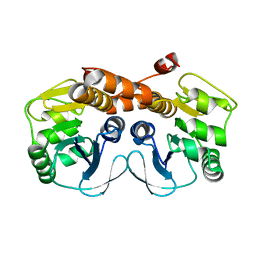 | | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum ATCC 824 | | Descriptor: | Intracellular protease/amidase related enzyme (ThiJ family) | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum
To be Published
|
|
3M9L
 
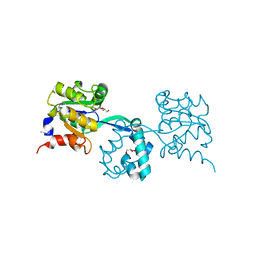 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 | | Descriptor: | GLYCEROL, Hydrolase, haloacid dehalogenase-like family | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-22 | | Release date: | 2010-04-07 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Had Family Hydrolase from Pseudomonas Fluorescens Pf-5
To be Published
|
|
2OO2
 
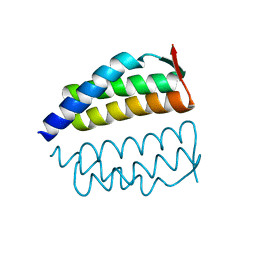 | | Crystal structure of protein AF1782 from Archaeoglobus fulgidus, Pfam DUF357 | | Descriptor: | Hypothetical protein AF_1782 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Adams, J, Sridhar, V, Smyth, L, Freeman, J, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the hypothetical AF_1782 protein from Archaeoglobus fulgidus
To be Published
|
|
2Q01
 
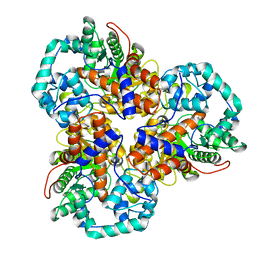 | | Crystal structure of glucuronate isomerase from Caulobacter crescentus | | Descriptor: | POTASSIUM ION, Uronate isomerase | | Authors: | Patskovsky, Y, Bonanno, J, Sridhar, V, Sauder, J.M, Freeman, J, Powell, A, Koss, J, Groshong, C, Gheyi, T, Wasserman, S.R, Raushel, F, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-18 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of Glucuronate Isomerase from Caulobacter crescentus.
To be Published
|
|
2QWT
 
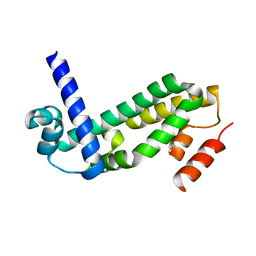 | | Crystal structure of the TetR transcription regulatory protein from Mycobacterium vanbaalenii | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Adams, J, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the TetR transcription regulatory protein from Mycobacterium vanbaalenii.
To be Published
|
|
2R5X
 
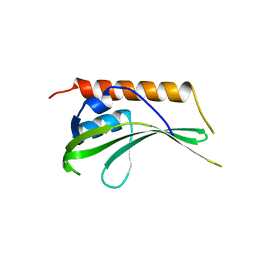 | | Crystal structure of uncharacterized conserved protein YugN from Geobacillus kaustophilus HTA426 | | Descriptor: | Uncharacterized conserved protein | | Authors: | Ramagopal, U.A, Patskovsky, Y, Shi, W, Toro, R, Meyer, A.J, Freeman, J, Wu, B, Koss, J, Groshong, C, Rodgers, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of uncharacterized protein YugN from Geobacillus kaustophilus HTA426.
To be Published
|
|
2OZ8
 
 | | Crystal structure of putative mandelate racemase from Mesorhizobium loti | | Descriptor: | Mll7089 protein, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-25 | | Release date: | 2007-03-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of putative mandelate racemase from Mesorhizobium loti
To be Published
|
|
2OX4
 
 | | Crystal structure of putative dehydratase from Zymomonas mobilis ZM4 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Dehydratase from Zymomonas Mobilis Zm4
To be Published
|
|
2OZ3
 
 | | Crystal structure of L-Rhamnonate dehydratase from Azotobacter vinelandii | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wu, B, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Rhamnonate dehydratase from azotobacter vinelandii
To be Published
|
|
2OO6
 
 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 | | Descriptor: | Putative L-alanine-DL-glutamate epimerase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Wu, B, Sridhar, V, Freeman, J, Smyth, L, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400
To be Published
|
|
2OO3
 
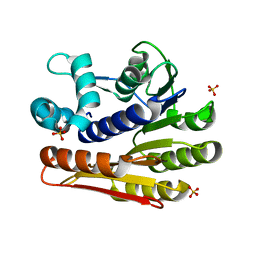 | | Crystal structure of protein LPL1258 from Legionella pneumophila str. Philadelphia 1, Pfam DUF519 | | Descriptor: | Protein involved in catabolism of external DNA, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Slocombe, A, Reyes, C, Ozyurt, S, Smyth, L, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hypothetical lpl1258 protein from Legionella pneumophila
To be Published
|
|
3I6T
 
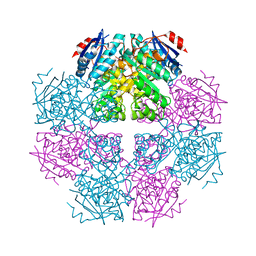 | | Crystal structure of muconate cycloisomerase from Jannaschia sp. | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, POTASSIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of muconate cycloisomerase from Jannaschia sp.
To be Published
|
|
2P61
 
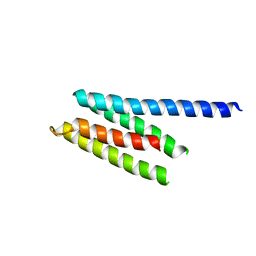 | | Crystal structure of protein TM1646 from Thermotoga maritima, Pfam DUF327 | | Descriptor: | Hypothetical protein TM_1646 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein TM_1646 from Thermotoga maritima
To be Published
|
|
2P1G
 
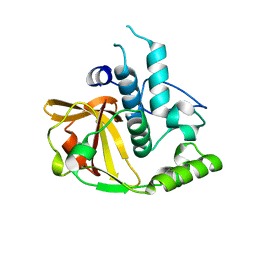 | | Crystal structure of a putative xylanase from Bacteroides fragilis | | Descriptor: | Putative xylanase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative xylanase from Bacteroides fragilis
To be Published
|
|
