2LJA
 
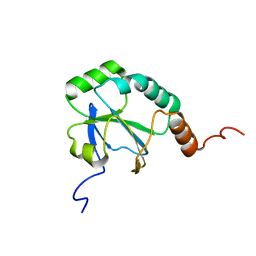 | | Solution Structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus | | Descriptor: | Putative thiol-disulfide oxidoreductase | | Authors: | Harris, R, Seidel, R.D, Hillerich, B, Gizzi, A, Kar, A, Lafleur, J, Chamala, S, Foti, R, Villegas, G, Evans, B, Zencheck, W, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-09-09 | | Release date: | 2011-10-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus
To be Published
|
|
2KUC
 
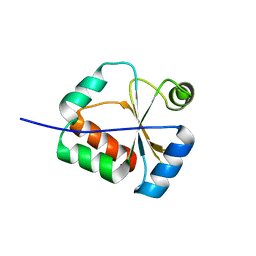 | | Solution Structure of a putative disulphide-isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Putative disulphide-isomerase | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-02 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a putative disulphide-isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
2L5L
 
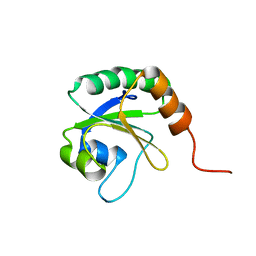 | | Solution Structure of Thioredoxin from Bacteroides Vulgatus | | Descriptor: | Thioredoxin | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-02 | | Release date: | 2010-11-24 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Thioredoxin from Bacteroides Vulgatus
To be Published
|
|
2L5O
 
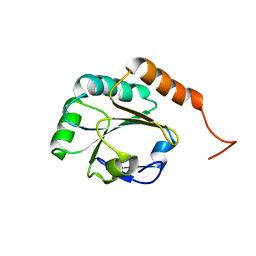 | | Solution Structure of a Putative Thioredoxin from Neisseria meningitidis | | Descriptor: | Putative thioredoxin | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-03 | | Release date: | 2010-12-15 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Putative Thioredoxin from Neisseria meningitidis
To be Published
|
|
2L57
 
 | | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens
To be Published
|
|
3ISA
 
 | | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase/isomerase FROM Bordetella parapertussis | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative enoyl-CoA hydratase/isomerase | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF enoyl-CoA hydratase FROM Bordetella parapertussis
To be Published
|
|
3IRV
 
 | | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae pv. phaseolicola 1448A | | Descriptor: | CYSTEINE HYDROLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae
To be Published
|
|
3IRS
 
 | | CRYSTAL STRUCTURE OF UNCHARACTERIZED TIM-BARREL PROTEIN BB4693 FROM Bordetella bronchiseptica | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Do, J, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF UNCHARACTERIZED HYDROLASE FROM Bordetella bronchiseptica
To be Published
|
|
3IVR
 
 | | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA ligase FROM Rhodopseudomonas palustris CGA009 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative long-chain-fatty-acid CoA ligase | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA SYNTHASE FROM Rhodopseudomonas palustris CGA009
To be Published
|
|
3JU2
 
 | | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein SMc04130 | | Authors: | Patskovsky, Y, Foti, R, Ramagopal, U, Malashkevich, V, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti
To be Published
|
|
3JVD
 
 | | Crystal structure of putative transcription regulation repressor (LACI family) FROM Corynebacterium glutamicum | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulators | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Foti, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-16 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LACI family protein from Corynebacterium glutamicum
To be Published
|
|
3RJL
 
 | | Crystal structure of 1-pyrroline-5-carboxylate dehydrogenase from Bacillus licheniformis (Target NYSGRC-000337) | | Descriptor: | 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, CADMIUM ION | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Seidel, R.D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-15 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of 1-Pyrroline-5-Carboxylate Dehydrogenase from Bacillus Licheniformis
To be Published
|
|
3RCY
 
 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme-like protein, ... | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035
To be Published
|
|
3P9X
 
 | | Crystal structure of phosphoribosylglycinamide formyltransferase from Bacillus Halodurans | | Descriptor: | GLYCEROL, SULFATE ION, phosphoribosylglycinamide formyltransferase | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Seidel, R.D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-10-18 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of phosphoribosylglycinamide formyltransferase from Bacillus Halodurans
To be Published
|
|
3QAN
 
 | | Crystal structure of 1-pyrroline-5-carboxylate dehydrogenase from bacillus halodurans | | Descriptor: | 1-pyrroline-5-carboxylate dehydrogenase 1, ACETATE ION | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Seidel, R.D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-01-11 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 1-Pyrroline-5-Carboxylate Dehydrogenase from Bacillus Halodurans
To be Published
|
|
3OA4
 
 | | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125 | | Descriptor: | GLYCEROL, SULFATE ION, ZINC ION, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Garrett, S, Foti, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125
To be Published
|
|
3OAJ
 
 | | CRYSTAL STRUCTURE OF putative dioxygenase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Putative ring-cleaving dioxygenase mhqO, SULFATE ION, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-08-05 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | CRYSTAL STRUCTURE OF putative dioxygenase from Bacillus subtilis subsp. subtilis str.
168
To be Published
|
|
3PQA
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Lactaldehyde dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661
To be Published
|
|
3PTW
 
 | | CRYSTAL STRUCTURE OF malonyl CoA-acyl carrier protein transacylase from Clostridium perfringens Atcc 13124 | | Descriptor: | Malonyl CoA-acyl carrier protein transacylase | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Seidel, R, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF malonyl CoA-acyl carrier protein transacylase from Clostridium perfringens
Atcc 13124
To be Published
|
|
3PRL
 
 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 | | Descriptor: | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-29 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125
To be Published
|
|
3RHD
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP | | Descriptor: | Lactaldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP
To be Published
|
|
4E5T
 
 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200750) from Labrenzia alexandrii DFL-11 | | Descriptor: | MAGNESIUM ION, Mandelate racemase / muconate lactonizing enzyme, C-terminal domain protein | | Authors: | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-14 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a putative MR/ML enzyme from Labrenzia alexandrii DFL-11
to be published
|
|
4E4U
 
 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200780) from Burkholderia SAR-1 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Mandalate racemase/muconate lactonizing enzyme | | Authors: | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-13 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a putative MR/ML enzyme from Burkholderia SAR-1
to be published
|
|
4E21
 
 | | The crystal structure of 6-phosphogluconate dehydrogenase from Geobacter metallireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 6-phosphogluconate dehydrogenase (Decarboxylating) | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-07 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | The crystal structure of 6-phosphogluconate dehydrogenase from Geobacter metallireducens
To be Published
|
|
2YB4
 
 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound SO4, no metal | | Descriptor: | AMIDOHYDROLASE, SULFATE ION | | Authors: | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-16 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
