4Z58
 
 | | HipB-O3 20mer complex | | 分子名称: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*TP*AP*CP*GP*GP*GP*AP*TP*AP*A)-3') | | 著者 | Min, J, Brennan, R.G, Schumacher, M.A. | | 登録日 | 2015-04-02 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular mechanism on hipBA gene regulation.
To be published
|
|
3UFD
 
 | | C.Esp1396I bound to its highest affinity operator site OM | | 分子名称: | CHLORIDE ION, DNA (5'-D(*AP*TP*GP*TP*AP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*TP*AP*CP*A)-3'), ... | | 著者 | Ball, N.J, McGeehan, J.E, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | 登録日 | 2011-11-01 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structural basis of differential DNA sequence recognition by restriction-modification controller proteins.
Nucleic Acids Res., 40, 2012
|
|
3VK0
 
 | | Crystal Structure of hypothetical transcription factor NHTF from Neisseria | | 分子名称: | Transcriptional regulator | | 著者 | Wang, H.-C, Ko, T.-P, Wu, M.-L, Wu, H.-J, Ku, S.-C, Wang, A.H.-J. | | 登録日 | 2011-11-01 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor
Nucleic Acids Res., 2012
|
|
2PPX
 
 | | Crystal structure of a HTH XRE-family like protein from Agrobacterium tumefaciens | | 分子名称: | GLYCEROL, SULFATE ION, Uncharacterized protein Atu1735 | | 著者 | Cuff, M.E, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-04-30 | | 公開日 | 2007-05-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a HTH XRE-family like protein from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
5A7L
 
 | |
2R1J
 
 | | Crystal Structure of the P22 c2 Repressor protein in complex with the synthetic operator 9T | | 分子名称: | 5'-D(*DCP*DAP*DTP*DTP*DTP*DAP*DAP*DGP*DAP*DTP*DAP*DTP*DCP*DTP*DTP*DAP*DAP*DAP*DTP*DA)-3', 5'-D(*DTP*DAP*DTP*DTP*DTP*DAP*DAP*DGP*DAP*DTP*DAP*DTP*DCP*DTP*DTP*DAP*DAP*DAP*DTP*DG)-3', Repressor protein C2 | | 著者 | Williams, L.D, Koudelka, G.B, Watkins, D, Hsiao, C, Woods, K. | | 登録日 | 2007-08-22 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | P22 c2 repressor-operator complex: mechanisms of direct and indirect readout
Biochemistry, 47, 2008
|
|
3QQ6
 
 | | The N-terminal DNA binding domain of SinR from Bacillus subtilis | | 分子名称: | HTH-type transcriptional regulator sinR | | 著者 | Colledge, V, Fogg, M.J, Levdikov, V.M, Dodson, E.J, Wilkinson, A.J. | | 登録日 | 2011-02-15 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and Organisation of SinR, the Master Regulator of Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 411, 2011
|
|
3ZHI
 
 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 | | 分子名称: | CI | | 著者 | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | 登録日 | 2012-12-21 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
2R63
 
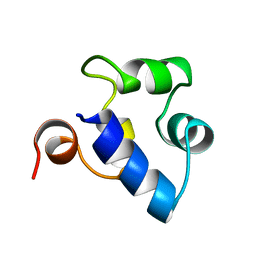 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | 著者 | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | 登録日 | 1996-11-13 | | 公開日 | 1997-06-16 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
3ZHM
 
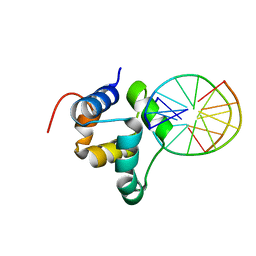 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 in complex with the OL2 operator half-site | | 分子名称: | 5'-D(*AP*CP*GP*TP*GP*AP*AP*CP*TP*TP*GP*CP*AP*CP *TP*TP*GP*A)-3', 5'-D(*AP*GP*TP*TP*CP*AP*CP*GP*TP*TP*CP*AP*AP*GP *TP*GP*CP*A)-3', CI | | 著者 | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | 登録日 | 2012-12-22 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
3S8Q
 
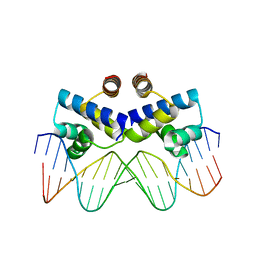 | | Crystal structure of the R-M controller protein C.Esp1396I OL operator complex | | 分子名称: | DNA (5'-D(*AP*TP*GP*TP*GP*AP*CP*TP*TP*AP*TP*AP*GP*TP*CP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*GP*GP*AP*CP*TP*AP*TP*AP*AP*GP*TP*CP*AP*CP*A)-3'), R-M CONTROLLER PROTEIN | | 著者 | McGeehan, J.E, Ball, N.J, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | 登録日 | 2011-05-30 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Recognition of dual symmetry by the controller protein C.Esp1396I based on the structure of the transcriptional activation complex.
Nucleic Acids Res., 40, 2012
|
|
3TRB
 
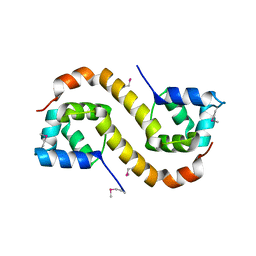 | | Structure of an addiction module antidote protein of a HigA (higA) family from Coxiella burnetii | | 分子名称: | Virulence-associated protein I | | 著者 | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | 登録日 | 2011-09-09 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3CEC
 
 | |
3CRO
 
 | |
1PER
 
 | |
3CLC
 
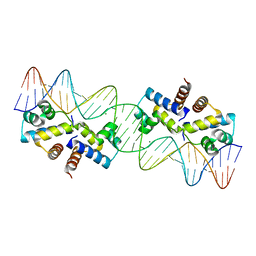 | | Crystal Structure of the Restriction-Modification Controller Protein C.Esp1396I Tetramer in Complex with its Natural 35 Base-Pair Operator | | 分子名称: | 35-MER, MAGNESIUM ION, Regulatory protein | | 著者 | McGeehan, J.E, Streeter, S.D, Thresh, S.J, Ball, N, Ravelli, R.B, Kneale, G.G. | | 登録日 | 2008-03-18 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural analysis of the genetic switch that regulates the expression of restriction-modification genes.
Nucleic Acids Res., 36, 2008
|
|
1PRA
 
 | |
6H49
 
 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | 分子名称: | Orf20, SULFATE ION | | 著者 | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | 登録日 | 2018-07-20 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
1R69
 
 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | REPRESSOR PROTEIN CI | | 著者 | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | 登録日 | 1988-12-08 | | 公開日 | 1989-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | 著者 | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | 登録日 | 1996-11-08 | | 公開日 | 1997-06-16 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
3EUS
 
 | |
1RPE
 
 | |
6IRP
 
 | |
7CSW
 
 | |
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | 分子名称: | HTH cro/C1-type domain-containing protein | | 著者 | Song, Y.J, Luo, G.H, Bao, R. | | 登録日 | 2020-08-17 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
