7UT1
 
 | |
7USF
 
 | |
7SEP
 
 | |
6U8Q
 
 | | CryoEM structure of HIV-1 cleaved synaptic complex (CSC) intasome | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-09-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
7SJX
 
 | | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | | Descriptor: | Gag-Pol polyprotein | | Authors: | Lyumkis, D, Passos, D, Arnold, E, Harrison, J.J.E.K, Ruiz, F.X. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Sci Adv, 8, 2022
|
|
6V3K
 
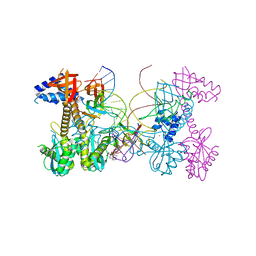 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ419 (compound 4c) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-(5-oxidanylpentyl)-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-11-25 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6VDK
 
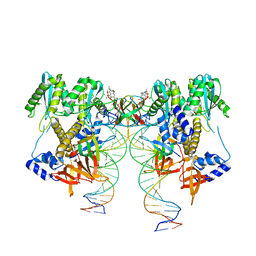 | | CryoEM structure of HIV-1 conserved Intasome Core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
6VOY
 
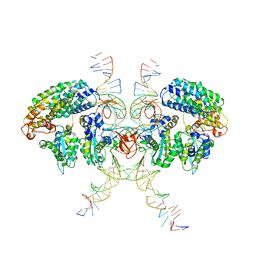 | | Cryo-EM structure of HTLV-1 instasome | | Descriptor: | DNA (25-MER), DNA (5'-D(P*AP*CP*AP*CP*AP*CP*TP*TP*GP*AP*CP*TP*AP*GP*GP*GP*TP*G)-3'), DNA-binding protein 7d, ... | | Authors: | Bhatt, V, Shi, K, Sundborger, A, Aihara, H. | | Deposit date: | 2020-02-01 | | Release date: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of host protein hijacking in human T-cell leukemia virus integration.
Nat Commun, 11, 2020
|
|
7Z0H
 
 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.6 A (focus subunit AC40). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-02-22 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
7Z2Z
 
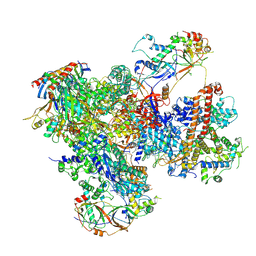 | | Structure of yeast RNA Polymerase III-DNA-Ty1 integrase complex (Pol III-DNA-IN1) at 3.1 A | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Nguyen, P.Q, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
7Z30
 
 | |
7Z31
 
 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.7 A (focus subunit C11, no C11 C-terminal Zn-ribbon in the funnel pore). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
5EJK
 
 | | Crystal structure of the Rous sarcoma virus intasome | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*T)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*CP*TP*C)-3'), ... | | Authors: | Yin, Z, Shi, K, Banerjee, S, Aihara, H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the Rous sarcoma virus intasome.
Nature, 530, 2016
|
|
5HOT
 
 | | Structural Basis for Inhibitor-Induced Aggregation of HIV-1 Integrase | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, Integrase | | Authors: | Gupta, K, Turkki, V, Sherrill-Mix, S, Hwang, Y, Eilers, G, Taylor, L, McDanal, C, Wang, P, Temelkoff, D, Nolte, R, Velthuisen, E, Jeffrey, J, Van Duyne, G.D, Bushman, F.D. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
6PUW
 
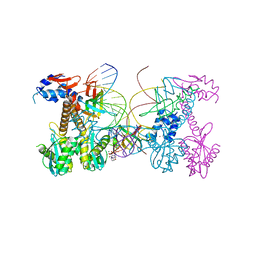 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and Bictegravir (BIC) | | Descriptor: | Bictegravir, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUZ
 
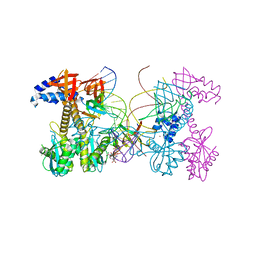 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUY
 
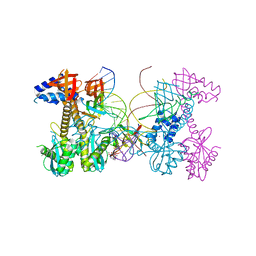 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ426 (compound 4d) | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUT
 
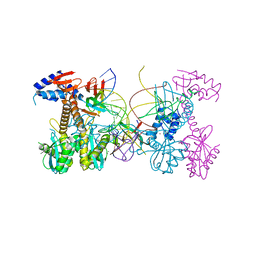 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with calcium | | Descriptor: | CALCIUM ION, Chimeric Sso7d and HIV-1 integrase, ZINC ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6RWO
 
 | | SIVrcm intasome (Q148H/G140S) in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWL
 
 | | SIVrcm intasome | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*AP*AP*GP*AP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*CP*CP*A)-3'), Pol protein, ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWN
 
 | | SIVrcm intasome in complex with dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWM
 
 | | SIVrcm intasome in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RNY
 
 | | PFV intasome - nucleosome strand transfer complex | | Descriptor: | DNA (108-MER), DNA (128-MER), DNA (33-MER), ... | | Authors: | Pye, V.E, Renault, L, Maskell, D.P, Cherepanov, P, Costa, A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
8E14
 
 | | Cryo-EM structure of Rous sarcoma virus strand transfer complex | | Descriptor: | DNA (42-MER), DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*TP*CP*TP*TP*CP*TP*TP*TP*C)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular determinants for Rous sarcoma virus intasome assemblies involved in retroviral integration.
J.Biol.Chem., 299, 2023
|
|
8FNH
 
 | | Structure of Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
