3SH9
 
 | | Crystal structure of fluorophore-labeled beta-lactamase PenP in complex with cefotaxime | | Descriptor: | 1-[6-(dimethylamino)naphthalen-2-yl]ethanone, Beta-lactamase, CEFOTAXIME, ... | | Authors: | Wong, W.-T, Zhao, Y.-X, Leung, Y.-C. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increased structural flexibility at the active site of a fluorophore-conjugated beta-lactamase distinctively impacts its binding toward diverse cephalosporin antibiotics
J.Biol.Chem., 286, 2011
|
|
3SOI
 
 | | Crystallographic structure of Bacillus licheniformis beta-lactamase W210F/W229F/W251F at 1.73 angstrom resolution | | Descriptor: | Beta-lactamase, CITRIC ACID | | Authors: | Acierno, J.P, Capaldi, S, Risso, V.A, Monaco, H.L, Ermacora, M.R. | | Deposit date: | 2011-06-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | X-ray evidence of a native state with increased compactness populated by tryptophan-less B. licheniformis beta-lactamase.
Protein Sci., 21, 2012
|
|
3TG9
 
 | | The crystal structure of penicillin binding protein from Bacillus halodurans | | Descriptor: | Penicillin-binding protein | | Authors: | Zhang, Z, Satyanarayana, L, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of penicillin binding protein from Bacillus halodurans
To be Published
|
|
3TOI
 
 | |
3TSG
 
 | | Crystal structure of GES-14 | | Descriptor: | Extended-spectrum beta-lactamase GES-14, GLYCEROL, IODIDE ION | | Authors: | Delbruck, H, Hoffmann, K.M.V, Bebrone, C. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and crystallographic studies of extended-spectrum GES-11, GES-12, and GES-14 beta-lactamases.
Antimicrob.Agents Chemother., 56, 2012
|
|
3V3R
 
 | | Crystal Structure of GES-11 | | Descriptor: | Extended spectrum class A beta-lactamase GES-11, IODIDE ION, SODIUM ION | | Authors: | Delbruck, H, Hoffmann, K.M.V, Bebrone, C. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Kinetic and crystallographic studies of extended-spectrum GES-11, GES-12, and GES-14 beta-lactamases.
Antimicrob.Agents Chemother., 56, 2012
|
|
3V3S
 
 | | Crystal structure of GES-18 | | Descriptor: | Extended spectrum beta-lactamase GES-18 | | Authors: | Delbruck, H, Hoffmann, K.M.V, Bebrone, C. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GES-18, a new carbapenem-hydrolyzing GES-Type beta-lactamase from pseudomonas aeruginosa that contains Ile80 and Ser170 residues.
Antimicrob.Agents Chemother., 57, 2013
|
|
3V50
 
 | | Complex of SHV S130G mutant beta-lactamase complexed to SA2-13 | | Descriptor: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, Beta-lactamase, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Wei, K, van den Akker, F. | | Deposit date: | 2011-12-15 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The importance of the trans-enamine intermediate as a beta-lactamase inhibition strategy probed in inhibitor-resistant SHV beta-lactamase variants.
Chemmedchem, 7, 2012
|
|
3V5M
 
 | |
3VFF
 
 | |
3VFH
 
 | |
3VWL
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187S/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWM
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187A/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWN
 
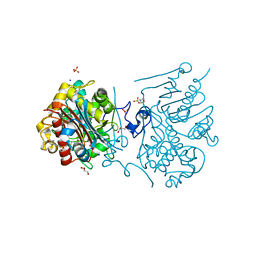 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187G/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWP
 
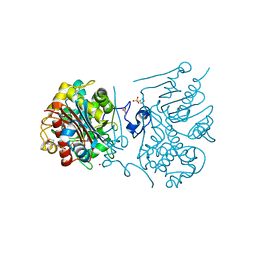 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWQ
 
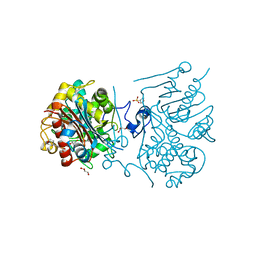 | | 6-aminohexanoate-dimer hydrolase S112A/G181D/R187A/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWR
 
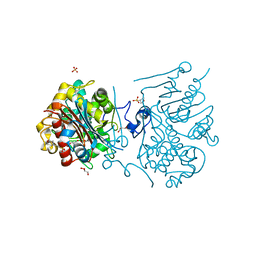 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187G/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3W4O
 
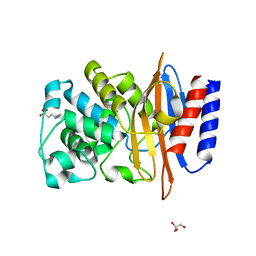 | | Crystal structure of PenI beta-lactamase from Burkholderia pseudomallei at pH9.5 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-lactamase, GLYCEROL | | Authors: | Nukaga, M, Ohuchi, N, Papp-Wallace, K.M, Taracila, M.A, Bonomo, R.A. | | Deposit date: | 2013-01-10 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants
J.Biol.Chem., 288, 2013
|
|
3W4P
 
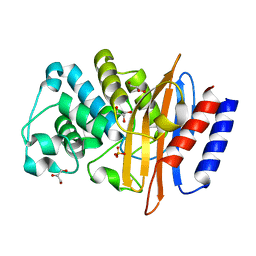 | | Crystal structure of PenI beta-lactamase from Burkholderia pseudomallei at pH7.5 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Nukaga, M, Ohuchi, N, Papp-Wallace, K.M, Taracila, M.A, Bonomo, R.A. | | Deposit date: | 2013-01-10 | | Release date: | 2013-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants
J.Biol.Chem., 288, 2013
|
|
3W4Q
 
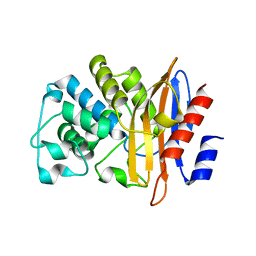 | | Crystal structure of PenA beta-lactamase from Burkholderia multivorans at pH4.2 | | Descriptor: | Beta-lactamase | | Authors: | Nukaga, M, Ohuchi, N, Papp-Wallace, K.M, Taracila, M.A, Bonomo, R.A. | | Deposit date: | 2013-01-10 | | Release date: | 2013-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants
J.Biol.Chem., 288, 2013
|
|
3W8K
 
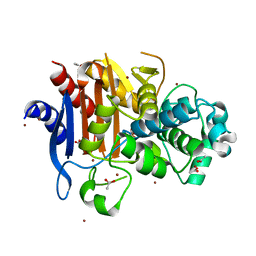 | | Crystal structure of class C beta-lactamase Mox-1 | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Shimizu-ibuka, A, Oguri, T, Furuyama, T, Ishii, Y. | | Deposit date: | 2013-03-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Mox-1, a unique plasmid-mediated class C beta-lactamase with hydrolytic activity towards moxalactam
Antimicrob.Agents Chemother., 58, 2014
|
|
3WRT
 
 | | Wild type beta-lactamase DERIVED FROM CHROMOHALOBACTER SP.560 | | Descriptor: | Beta-lactamase | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRZ
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (without soaking) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS0
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1A) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS1
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
