3VZQ
 
 | |
3VZP
 
 | | Crystal structure of PhaB from Ralstonia eutropha | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Acetoacetyl-CoA reductase, GLYCEROL, ... | | Authors: | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
3VZS
 
 | | Crystal structure of PhaB from Ralstonia eutropha in complex with Acetoacetyl-CoA and NADP | | Descriptor: | ACETOACETYL-COENZYME A, Acetoacetyl-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
4HBG
 
 | |
4N5L
 
 | |
4N5M
 
 | |
4N5N
 
 | |
4O5O
 
 | |
4J7X
 
 | |
4C7K
 
 | | 11b-Hydroxysteroid Dehydrogenase Type I in complex with inhibitor | | Descriptor: | 2-ethyl-N-[(1S,3R)-5-oxidanyl-2-adamantyl]-4-[(2R)-oxolan-2-yl]-1,3-thiazole-5-carboxamide, CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Goldberg, F.W, Dossetter, A.G, Scott, J.S, Robb, G.R, Boyd, S, Groombridge, S.D, Kemmitt, P.D, Sjogren, T, Morentin Gutierrez, P, de Schoolmeester, J, Swales, J.G, Turnbull, A.V, Wild, M.J. | | Deposit date: | 2013-09-23 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Optimization of Brain Penetrant 11Beta-Hydroxysteroid Dehydrogenase Type I Inhibitors and in Vivo Testing in Diet- Induced Obese Mice.
J.Med.Chem., 57, 2014
|
|
3W8D
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor methylmalonate | | Descriptor: | CHLORIDE ION, D-3-hydroxybutyrate dehydrogenase, METHYLMALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray diffraction of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and methylmalonate
To be Published
|
|
3W8E
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and a substrate D-3-hydroxybutyrate | | Descriptor: | (3R)-3-hydroxybutanoic acid, D-3-hydroxybutyrate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | High resolution X-ray diffraction of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and D-3-hydroxybutyrate
To be Published
|
|
3W8F
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor malonate | | Descriptor: | CHLORIDE ION, D-3-hydroxybutyrate dehydrogenase, MALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray diffraction of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and malonate
To be Published
|
|
4BMV
 
 | | Short-chain dehydrogenase from Sphingobium yanoikuyae in complex with NADPH | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SHORT-CHAIN DEHYDROGENASE | | Authors: | Man, H, Kedziora, K, Lavandera-Garcia, I, Gotor-Fernandez, V, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
4NMH
 
 | | 11-beta-HSD1 in complex with a 3,3-Di-methyl-azetidin-2-one | | Descriptor: | (4S)-4-(2-methoxyphenyl)-3,3-dimethyl-1-[3-(methylsulfonyl)phenyl]azetidin-2-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McCoull, W, Augustin, M, Blake, C, Ertan, A, Kilgour, E.K, Krapp, S, Moore, J.E, Newcombe, N.J, Packer, M.J, Rees, A, Revill, J, Scott, J.S, Selmi, N, Gerhardt, S, Ogg, D.J, Steinbacher, S, Whittamore, P.R.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification and optimisation of 3,3-dimethyl-azetidin-2-ones as potent and selective inhibitors of 11 beta-hydroxysteroid dehydrogenase type 1 (11-beta-HSD1)
TO BE PUBLISHED
|
|
4K1L
 
 | | 4,4-Dioxo-5,6-dihydro-[1,4,3]oxathiazines, a novel class of 11 beta-HSD1 inhibitors for the treatment of diabetes | | Descriptor: | (4aS,8aR)-N-cyclohexyl-4a,5,6,7,8,8a-hexahydro-4,1,2-benzoxathiazin-3-amine 1,1-dioxide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Von der Heyden, C, Engel, C.K. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 1,1-Dioxo-5,6-dihydro-[4,1,2]oxathiazines, a novel class of 11-HSD1 inhibitors for the treatment of diabetes.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4K26
 
 | | 4,4-Dioxo-5,6-dihydro-[1,4,3]oxathiazines, a novel class of 11 -HSD1 inhibitors for the treatment of diabetes | | Descriptor: | (4aS,8aR)-N-cyclohexyl-4a,5,6,7,8,8a-hexahydro-4,1,2-benzoxathiazin-3-amine 1,1-dioxide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Loenze, P, Schimanski-Breves, S, Vonderheyden, C, Engel, C.K. | | Deposit date: | 2013-04-08 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 1,1-Dioxo-5,6-dihydro-[4,1,2]oxathiazines, a novel class of 11-HSD1 inhibitors for the treatment of diabetes.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4P38
 
 | | Human 11beta-Hydroxysteroid Dehydrogenase Type 1 in complex with AZD8329 | | Descriptor: | 4-[4-(2-adamantylcarbamoyl)-5-tert-butyl-pyrazol-1-yl]benzoic acid, CHLORIDE ION, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Ogg, D, Hargreaves, D, Gerhardt, S. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel acidic 11 beta-hydroxysteroid dehydrogenase type 1 (11 beta-HSD1) inhibitor with reduced acyl glucuronide liability: the discovery of 4-[4-(2-adamantylcarbamoyl)-5-tert-butyl-pyrazol-1-yl]benzoic acid (AZD8329).
J.Med.Chem., 55, 2012
|
|
4IJW
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with 3-[1-(4-chlorophenyl)cyclopropyl]-8-cyclopropyl[1,2,4]triazolo[4,3-a]pyridine | | Descriptor: | 3-[1-(4-chlorophenyl)cyclopropyl]-8-cyclopropyl[1,2,4]triazolo[4,3-a]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
4IJU
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with (1S,4S)-4-[8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridin-3-yl]bicyclo[2.2.1]heptan-1-ol | | Descriptor: | (1s,4s)-4-[8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridin-3-yl]bicyclo[2.2.1]heptan-1-ol, CHLORIDE ION, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
4IJV
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with 3-[1-(4-chlorophenyl)cyclopropyl]-8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridine | | Descriptor: | 3-[1-(4-chlorophenyl)cyclopropyl]-8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
4CR7
 
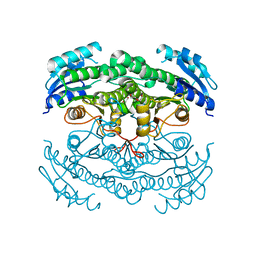 | | Crystal structure of the N-acetyl-D-mannosamine dehydrogenase with n-acetylmannosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, N-ACYLMANNOSAMINE 1-DEHYDROGENASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Gil-Ortiz, F, Sola-Carvajal, A, Garcia-Carmona, F, Sanchez-Ferrer, A, Rubio, V. | | Deposit date: | 2014-02-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures and Functional Studies Clarify Substrate Selectivity and Catalytic Residues for the Unique Orphan Enzyme N-Acetyl-D-Mannosamine Dehydrogenase.
Biochem.J., 462, 2014
|
|
4CR8
 
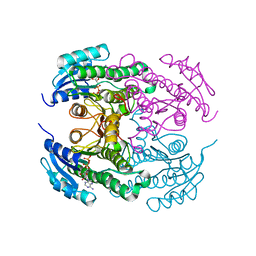 | | Crystal structure of the N-acetyl-D-mannosamine dehydrogenase with NAD | | Descriptor: | N-ACYLMANNOSAMINE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gil-Ortiz, F, Sola-Carvajal, A, Garcia-Carmona, F, Sanchez-Ferrer, A, Rubio, V. | | Deposit date: | 2014-02-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures and Functional Studies Clarify Substrate Selectivity and Catalytic Residues for the Unique Orphan Enzyme N-Acetyl-D-Mannosamine Dehydrogenase.
Biochem.J., 462, 2014
|
|
4CR6
 
 | | Crystal structure of the N-acetyl-D-mannosamine dehydrogenase without substrates | | Descriptor: | N-ACYLMANNOSAMINE 1-DEHYDROGENASE, alpha-D-mannopyranose | | Authors: | Gil-Ortiz, F, Sola-Carvajal, A, Garcia-Carmona, F, Sanchez-Ferrer, A, Rubio, V. | | Deposit date: | 2014-02-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures and Functional Studies Clarify Substrate Selectivity and Catalytic Residues for the Unique Orphan Enzyme N-Acetyl-D-Mannosamine Dehydrogenase.
Biochem.J., 462, 2014
|
|
4TRR
 
 | |
