8AD4
 
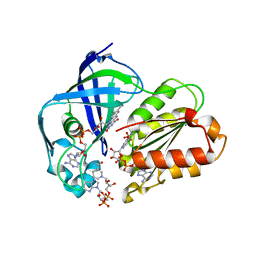 | | X-ray structure of NqrF(129-408)of Vibrio cholerae in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AD5
 
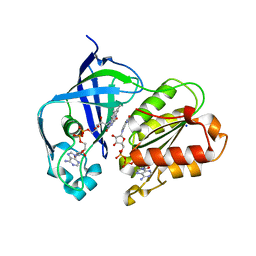 | | X-ray structure of NqrF(129-408)of Vibrio cholerae variant F406A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7S24
 
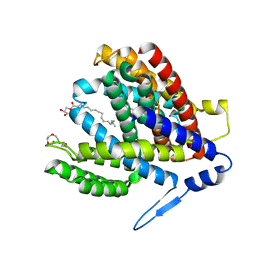 | | Crystal structure of the Na+/H+ antiporter NhaA at pH 6.5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Na(+)/H(+) antiporter NhaA, PENTAETHYLENE GLYCOL | | Authors: | Drew, D, Brock, J, Uzdavinys, P, Matsuoka, R. | | Deposit date: | 2021-09-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Na + /H + antiporter NhaA at active pH reveals the mechanistic basis for pH sensing.
Nat Commun, 13, 2022
|
|
7T9G
 
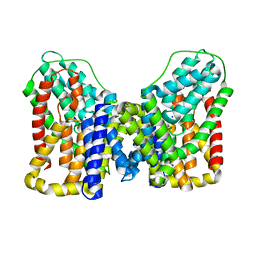 | | Structure of VcINDY-Na+ | | Descriptor: | DASS family sodium-coupled anion symporter, SODIUM ION | | Authors: | Sauer, D.B, Marden, J.J, Song, J.M, Wang, D.N. | | Deposit date: | 2021-12-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis of ion - substrate coupling in the Na + -dependent dicarboxylate transporter VcINDY.
Nat Commun, 13, 2022
|
|
7WYT
 
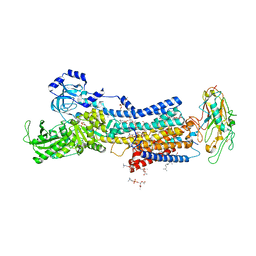 | | Crystal structures of Na+,K+-ATPase in complex with ouabain | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WYS
 
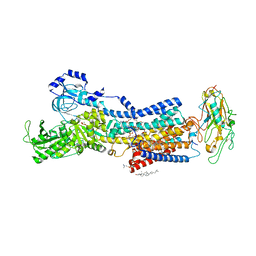 | | Crystal structures of Na+,K+-ATPase in complex with istaroxime | | Descriptor: | (3E,5S,8R,9S,10R,13S,14S)-3-(2-azanylethoxyimino)-10,13-dimethyl-1,2,4,5,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthrene-6,17-dione, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WYY
 
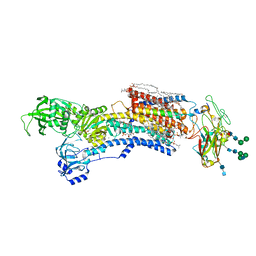 | | Cryo-EM structure of Na+,K+-ATPase in the E2P state formed by inorganic phosphate with istaroxime | | Descriptor: | (3E,5S,8R,9S,10R,13S,14S)-3-(2-azanylethoxyimino)-10,13-dimethyl-1,2,4,5,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthrene-6,17-dione, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WZ0
 
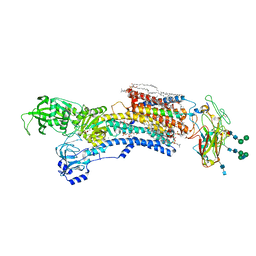 | | Cryo-EM structure of Na+,K+-ATPase in the E2P state formed by inorganic phosphate with ouabain | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WYW
 
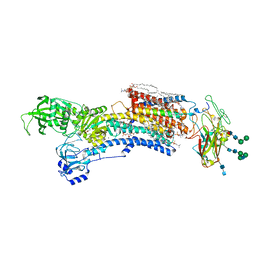 | | Cryo-EM structure of Na+,K+-ATPase in the E2P state formed by inorganic phosphate | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2HTG
 
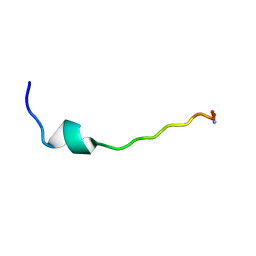 | | Structural and functional characterization of TM VII of the NHE1 isoform of the Na+/H+ exchanger | | Descriptor: | NHE1 isoform of Na+/H+ exchanger 1 | | Authors: | Rainey, J.K, Ding, J, Xu, C, Fliegel, L, Sykes, B.D. | | Deposit date: | 2006-07-25 | | Release date: | 2006-08-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of transmembrane segment VII of the Na(+)/H(+) exchanger isoform 1
J.Biol.Chem., 281, 2006
|
|
4RET
 
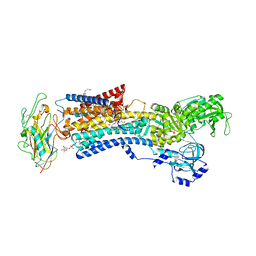 | | Crystal structure of the Na,K-ATPase E2P-digoxin complex with bound magnesium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Gregersen, J.L, Laursen, M, Yatime, L, Nissen, P, Fedosova, N.U. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RES
 
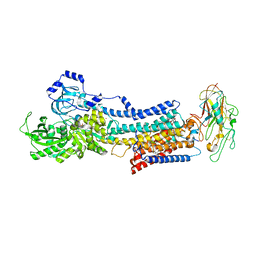 | | Crystal structure of the Na,K-ATPase E2P-bufalin complex with bound potassium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Laursen, M, Yatime, L, Gregersen, J.L, Nissen, P, Fedosova, N.U. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3JZ1
 
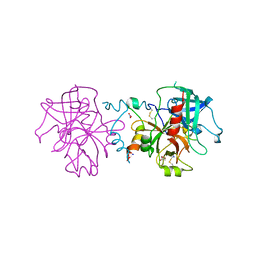 | | Crystal structure of human thrombin mutant N143P in E:Na+ form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NITRATE ION, ... | | Authors: | Niu, W, Chen, Z, Bush-Pelc, L.A, Bah, A, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-09-22 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutant N143P reveals how Na+ activates thrombin
J.Biol.Chem., 284, 2009
|
|
2WGM
 
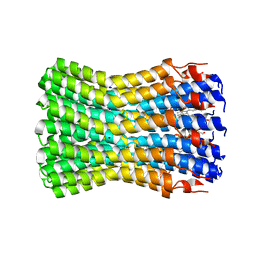 | | Complete ion-coordination structure in the rotor ring of Na-dependent F-ATP synthase | | Descriptor: | ATP SYNTHASE SUBUNIT C, SODIUM ION SPECIFIC, NONAN-1-OL, ... | | Authors: | Meier, T, Pogoryelov, D, Diederichs, K. | | Deposit date: | 2009-04-21 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Complete Ion-Coordination Structure in the Rotor Ring of Na(+)-Dependent F-ATP Synthases.
J.Mol.Biol., 391, 2009
|
|
3E6P
 
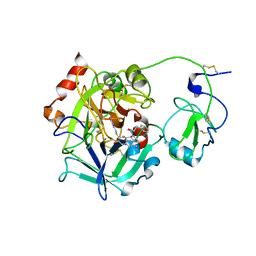 | | Crystal structure of human meizothrombin desF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-PHENYLALANYL-N-[(1S)-4-{[(Z)-AMINO(IMINO)METHYL]AMINO}-1-(CHLOROACETYL)BUTYL]-L-PROLINAMIDE, Prothrombin, ... | | Authors: | Papaconstantinou, M.E, Gandhi, P, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2008-08-15 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Na(+) binding to meizothrombin desF1.
Cell.Mol.Life Sci., 65, 2008
|
|
2A2Q
 
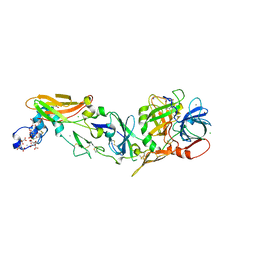 | | Complex of Active-site Inhibited Human Coagulation Factor VIIa with Human Soluble Tissue Factor in the Presence of Ca2+, Mg2+, Na+, and Zn2+ | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII, ... | | Authors: | Bajaj, S.P, Bajaj, M, Schmidt, A.E, Padmanabhan, K. | | Deposit date: | 2005-06-22 | | Release date: | 2006-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of p-aminobenzamidine- and benzamidine-VIIa/soluble tissue factor: unpredicted conformation of the 192-193 peptide bond and mapping of Ca2+, Mg2+, Na+, and Zn2+ sites in factor VIIa.
J.Biol.Chem., 281, 2006
|
|
1BG5
 
 | | CRYSTAL STRUCTURE OF THE ANKYRIN BINDING DOMAIN OF ALPHA-NA,K-ATPASE AS A FUSION PROTEIN WITH GLUTATHIONE S-TRANSFERASE | | Descriptor: | FUSION PROTEIN OF ALPHA-NA,K-ATPASE WITH GLUTATHIONE S-TRANSFERASE | | Authors: | Zhang, Z, Devarajan, P, Morrow, J.S. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the ankyrin-binding domain of alpha-Na,K-ATPase.
J.Biol.Chem., 273, 1998
|
|
3E9U
 
 | | Crystal structure of Calx CBD2 domain | | Descriptor: | Na/Ca exchange protein | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2008-08-23 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CBD2 from the Drosophila Na(+)/Ca(2+) exchanger: diversity of Ca(2+) regulation and its alternative splicing modification.
J.Mol.Biol., 387, 2009
|
|
1LPI
 
 | | HEW LYSOZYME: TRP...NA CATION-PI INTERACTION | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Wouters, J. | | Deposit date: | 1998-04-15 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cation-pi (Na+-Trp) interactions in the crystal structure of tetragonal lysozyme.
Protein Sci., 7, 1998
|
|
1Q3I
 
 | | Crystal Structure of Na,K-ATPase N-domain | | Descriptor: | NICKEL (II) ION, Na,K-ATPase | | Authors: | Hakansson, K.O. | | Deposit date: | 2003-07-30 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystallographic Structure of Na,K-ATPase N-domain at 2.6 A Resolution
J.Mol.Biol., 332, 2003
|
|
4U9Q
 
 | | Crystal structure of NqrA in spacegroup P21 | | Descriptor: | Na(+)-translocating NADH-quinone reductase subunit A | | Authors: | Fritz, G. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
8JBM
 
 | | Crystal structure of Na+,K+-ATPase in the E1.Mn2+ state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JBL
 
 | | Crystal structure of Na+,K+-ATPase in the E1.Mg2+ state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
4XE5
 
 | | Crystal structure of the Na,K-ATPase from bovine | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, OUABAIN, ... | | Authors: | Gregersen, J.L, Mattle, D, Fedosova, N.U, Nissen, P, Reinhard, L. | | Deposit date: | 2014-12-22 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | Isolation, crystallization and crystal structure determination of bovine kidney Na(+),K(+)-ATPase.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5AW0
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 55 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
