7LW1
 
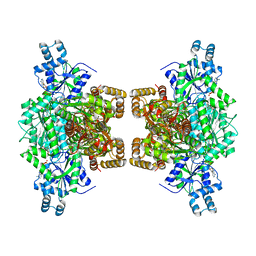 | | Human phosphofructokinase-1 liver type bound to activator NA-11 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B. | | Deposit date: | 2021-02-27 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Selective activation of PFKL suppresses the phagocytic oxidative burst.
Cell, 184, 2021
|
|
7UUZ
 
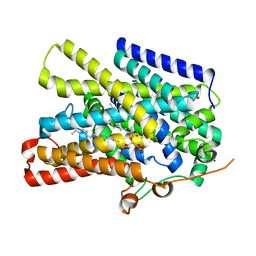 | | Structure of the sodium/iodide symporter (NIS) in complex with perrhenate and sodium | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, PERRHENATE, SODIUM ION, ... | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
7UV0
 
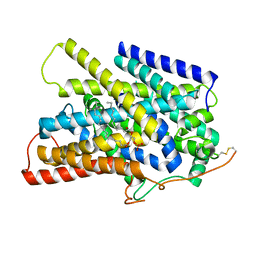 | | Structure of the sodium/iodide symporter (NIS) in complex with iodide and sodium | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, IODIDE ION, SODIUM ION, ... | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
6ZPL
 
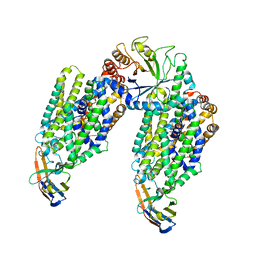 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
5HXZ
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
8D79
 
 | | Crystal structure of a four-tetrad, parallel, and Na+ stabilized Tetrahymena thermophila telomeric G-quadruplex in complex with N-methyl mesoporphyrin IX | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*T)-3'), N-METHYLMESOPORPHYRIN, ... | | Authors: | Chen, E.V, Beseiso, D, Yatsunyk, L.A. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Biophysical and structural characterization of telomeric G-quadruplexes from Tetrahymena thermophila in complex with N-Methyl Mesoporphyrin IX
To Be Published
|
|
7DJ2
 
 | | Crystal structure of the G26C/E290S mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DII
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 7 | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
8GNK
 
 | | CryoEM structure of cytosol-facing, substrate-bound ratGAT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Nayak, S.R, Joseph, D, Penmatsa, A. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of GABA transporter 1 reveals substrate recognition and transport mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6V64
 
 | | Crystal structure of human thrombin bound to ppack with tryptophans replaced by 5-F-tryptophan | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Ruben, E.A, Chen, Z, Di Cera, E. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.
J.Biol.Chem., 295, 2020
|
|
6O6J
 
 | |
2PGB
 
 | | Inhibitor-free human thrombin mutant C191A-C220A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, SULFATE ION | | Authors: | Bush-Pelc, L.A, Marino, F, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-04-09 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Important role of the cys-191 cys-220 disulfide bond in thrombin function and allostery
J.Biol.Chem., 282, 2007
|
|
6RFB
 
 | | Crystal structure of the potassium-pumping S254A mutant of the light-driven sodium pump KR2 in the monomeric form, pH 4.3 | | Descriptor: | EICOSANE, GLYCEROL, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6A90
 
 | | Complex of voltage-gated sodium channel NavPaS from American cockroach Periplaneta americana and Dc1a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mu-diguetoxin-Dc1a, ... | | Authors: | Shen, H.Z, li, Z.Q, Jiang, Y, Pan, X.J, Wu, J.P, Cristofori-Armstrong, B, Smith, J.J, Chin, Y.K.Y, Lei, J.L, Zhou, Q, King, G.F, Yan, N. | | Deposit date: | 2018-07-11 | | Release date: | 2018-08-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the modulation of voltage-gated sodium channels by animal toxins.
Science, 362, 2018
|
|
6A95
 
 | | Complex of voltage-gated sodium channel NavPaS from American cockroach Periplaneta americana bound with tetrodotoxin and Dc1a | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H.Z, li, Z.Q, Jiang, Y, Pan, X.J, Wu, J.P, Cristofori-Armstrong, B, Smith, J.J, Chin, Y.K.Y, Lei, J.L, Zhou, Q, King, G.F, Yan, N. | | Deposit date: | 2018-07-11 | | Release date: | 2018-08-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the modulation of voltage-gated sodium channels by animal toxins.
Science, 362, 2018
|
|
3TVB
 
 | | A Highly Symmetric DNA G-4 Quadruplex/drug Complex | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*GP*GP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Clark, G.R, Pytel, P.D, Squire, C.J. | | Deposit date: | 2011-09-19 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The high-resolution crystal structure of a parallel intermolecular DNA G-4 quadruplex/drug complex employing syn glycosyl linkages.
Nucleic Acids Res., 40, 2012
|
|
4M48
 
 | | X-ray structure of dopamine transporter elucidates antidepressant mechanism | | Descriptor: | 9D5 antibody, heavy chain, light chain, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | X-ray structure of dopamine transporter elucidates antidepressant mechanism.
Nature, 503, 2013
|
|
3KDP
 
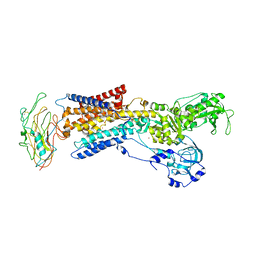 | | Crystal structure of the sodium-potassium pump | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, Na+/K+ ATPase gamma subunit transcript variant a, ... | | Authors: | Morth, J.P, Pedersen, B.P, Nissen, P. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the sodium-potassium pump.
Nature, 450, 2007
|
|
4WUB
 
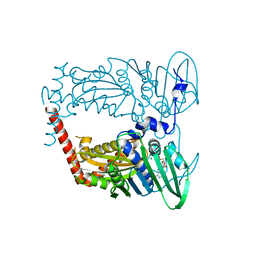 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WUD
 
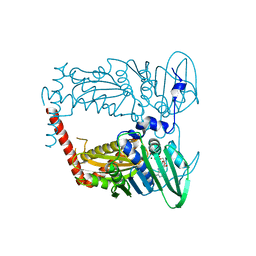 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from no salt condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3V5U
 
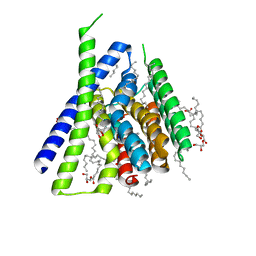 | | Structure of Sodium/Calcium Exchanger from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ACETATE ION, CALCIUM ION, ... | | Authors: | Jiang, Y, Liao, J, Li, H, Zeng, W, Sauer, D, Belmares, R. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger.
Science, 335, 2012
|
|
4DT7
 
 | | Crystal structure of thrombin bound to the activation domain QEDQVDPRLIDGKMTRRGDS of protein C | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Pozzi, N, Barranco-Medina, S, Chen, Z, Di Cera, E. | | Deposit date: | 2012-02-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposure of R169 controls protein C activation and autoactivation.
Blood, 120, 2012
|
|
4XTJ
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl plus 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YI9
 
 | |
3L8L
 
 | | Gramicidin D complex with sodium iodide | | Descriptor: | GRAMICIDIN D, IODIDE ION, METHANOL, ... | | Authors: | Olczak, A, Glowka, M.L, Szczesio, M, Bojarska, J, Wawrzak, Z, Duax, W.L. | | Deposit date: | 2009-12-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The first crystal structure of a gramicidin complex with sodium: high-resolution study of a nonstoichiometric gramicidin D-NaI complex.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
