4XA7
 
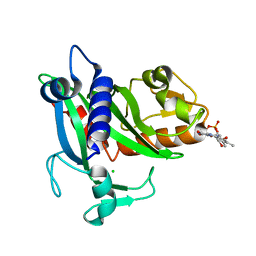 | | Soluble part of holo NqrC from V. harveyi | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit C | | Authors: | Borshchevskiy, V, Round, E, Bertsova, Y, Polovinkin, V, Gushchin, I, Mishin, A, Kovalev, K, Kachalova, G, Popov, A, Bogachev, A, Gordeliy, V. | | Deposit date: | 2014-12-12 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and Functional Investigation of Flavin Binding Center of the NqrC Subunit of Sodium-Translocating NADH:Quinone Oxidoreductase from Vibrio harveyi.
Plos One, 10, 2015
|
|
6EID
 
 | | Crystal structure of wild-type Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, PHOSPHATE ION, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
4MD2
 
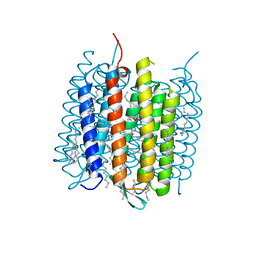 | | Ground state of bacteriorhodopsin from Halobacterium salinarum | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Borshchevskiy, V, Erofeev, I, Round, E, Weik, M, Ishchenko, A, Gushchin, I, Mishin, A, Bueldt, G, Gordeliy, V. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Low-dose X-ray radiation induces structural alterations in proteins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MD1
 
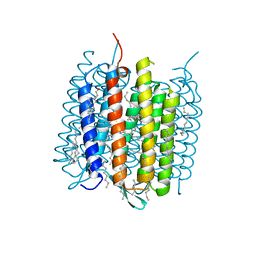 | | Orange species of bacteriorhodopsin from Halobacterium salinarum | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Borshchevskiy, V, Erofeev, I, Round, E, Weik, M, Ishchenko, A, Gushchin, I, Mishin, A, Bueldt, G, Gordeliy, V. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Low-dose X-ray radiation induces structural alterations in proteins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3MBV
 
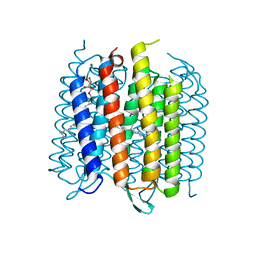 | | Structure of bacterirhodopsin crystallized in betta-XylOC(16+4) meso phase | | Descriptor: | (3R,7R,11R)-3,7,11,15-tetramethylhexadecyl alpha-D-ribopyranoside, Bacteriorhodopsin, RETINAL | | Authors: | Borshchevskiy, V, Moiseeva, E, Kuklin, A, Bueldt, G, Hato, M, Gordeliy, V. | | Deposit date: | 2010-03-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoprenoid-chained lipid beta-XylOC16+4 A novel molecule for in meso membrane protein crystallization
J.Cryst.Growth, 312, 2010
|
|
3NS0
 
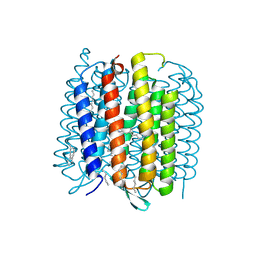 | | X-ray structure of bacteriorhodopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Borshchevskiy, V.I. | | Deposit date: | 2010-07-01 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray-Radiation-Induced Changes in Bacteriorhodopsin Structure.
J.Mol.Biol., 409, 2011
|
|
3NSB
 
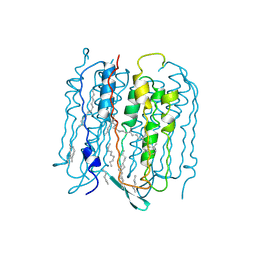 | |
7Z0D
 
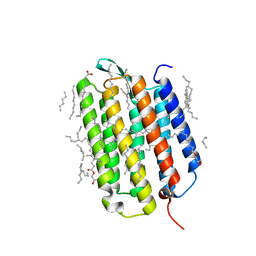 | | Crystal structure of the L state of bacteriorhodopsin at 1.20 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z09
 
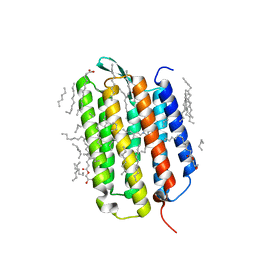 | | Crystal structure of the ground state of bacteriorhodopsin at 1.05 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0A
 
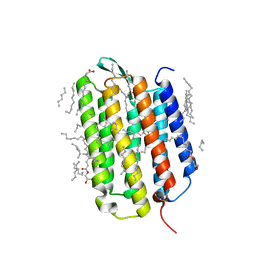 | | Crystal structure of the ground state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0E
 
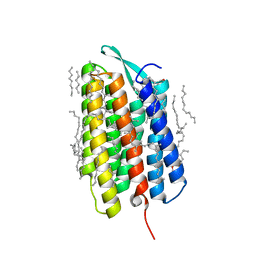 | | Crystal structure of the M state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0C
 
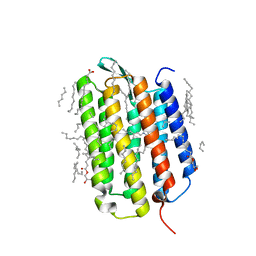 | | Crystal structure of the K state of bacteriorhodopsin at 1.53 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8RT2
 
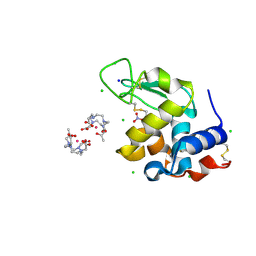 | | Hen Egg White Lysozyme Crystallized with Bioassembler on Earth | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | MacCarthy, C, Koudan, E.V, Petrov, S.V, Levin, A.A, Khesuani, Y.D, Borshchevskiy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Hen Egg White Lysozyme Crystallized with Bioassembler on Earth
To Be Published
|
|
8RT3
 
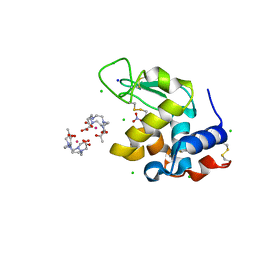 | | Hen Egg White Lysozyme Crystallized with Bioassembler in Space Microgravity Conditions | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | MacCarthy, C, Koudan, E.V, Petrov, S.V, Levin, A.A, Khesuani, Y.D, Borshchevskiy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Hen Egg White Lysozyme Crystallized with Bioassembler in Space Microgravity Conditions
To Be Published
|
|
8C6O
 
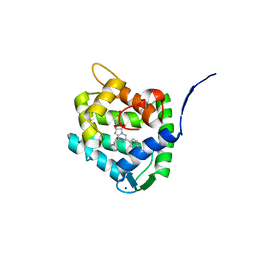 | | Crystal Structure of H64F obelin mutant from Obelia longissima at 2.2 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin, SODIUM ION | | Authors: | Natashin, P.V, Burakova, L.P, Kovaleva, M.I, Schevtsov, M.B, Dmitrieva, D.A, Eremeeva, E.V, Markova, S.V, Mishin, A.V, Borshchevskiy, V.I, Vysotski, E.S. | | Deposit date: | 2023-01-12 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of Tyr-His-Trp Triad and Water Molecule Near the N1-Atom of 2-Hydroperoxycoelenterazine in Bioluminescence of Hydromedusan Photoproteins: Structural and Mutagenesis Study.
Int J Mol Sci, 24, 2023
|
|
5VN9
 
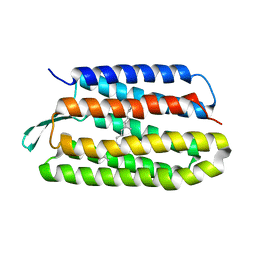 | | Structure of bacteriorhodopsin from crystals grown at 4 deg C using GlyNCOC15+4 as an LCP host lipid | | Descriptor: | Bacteriorhodopsin | | Authors: | Ishchenko, A, Peng, L, Zinovev, E, Vlasov, A, Lee, S.C, Kuklin, A, Mishin, A, Borshchevskiy, V, Zhang, Q, Cherezov, V. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Chemically Stable Lipids for Membrane Protein Crystallization.
Cryst Growth Des, 17, 2017
|
|
4HYJ
 
 | | Crystal structure of Exiguobacterium sibiricum rhodopsin | | Descriptor: | EICOSANE, RETINAL, Rhodopsin | | Authors: | Gushchin, I, Chervakov, P, Kuzmichev, P, Popov, A, Round, E, Borshchevskiy, V, Dolgikh, D, Kirpichnikov, M, Petrovskaya, L, Chupin, V, Arseniev, A, Gordeliy, V. | | Deposit date: | 2012-11-13 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5VN7
 
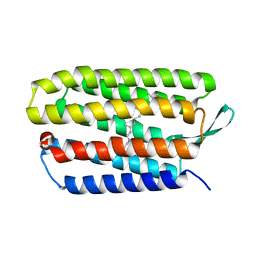 | | Structure of bacteriorhodopsin from crystals grown at 20 deg Celcius using GlyNCOC15+4 as an LCP host lipid | | Descriptor: | Bacteriorhodopsin | | Authors: | Ishchenko, A, Peng, L, Zinovev, E, Vlasov, A, Lee, S.C, Kuklin, A, Mishin, A, Borshchevskiy, V, Zhang, Q, Cherezov, V. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chemically Stable Lipids for Membrane Protein Crystallization.
Cryst Growth Des, 17, 2017
|
|
7NUX
 
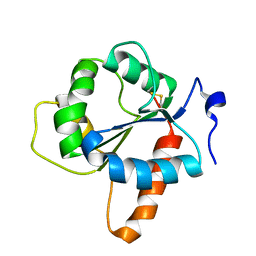 | | Crystal structure of the TIR domain of human TLR1 (crystallised without ZN2+ ions) | | Descriptor: | Toll-like receptor 1 | | Authors: | Vakhrameev, D.D, Luginina, A.P, Shevtsov, M.B, Lushpa, V.A, Mineev, K.S, Borshchevskiy, V.I. | | Deposit date: | 2021-03-15 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Modulation of Toll-like receptor 1 intracellular domain structure and activity by Zn 2+ ions.
Commun Biol, 4, 2021
|
|
8CIK
 
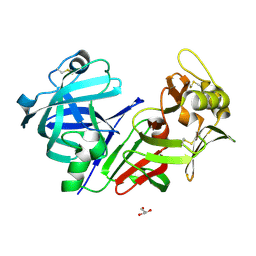 | | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution | | Descriptor: | Chymosin, GLYCEROL | | Authors: | Diusenova, S.E, Shevtsov, M.B, Borshchevskiy, V.I, Belenkaya, S.V, Kolybalov, D.S, Arkhipov, S.G, Volosnikova, E.A, Elchaninov, V.V, Shcherbakov, D.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution
To Be Published
|
|
7NUW
 
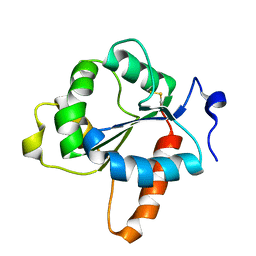 | | Crystal structure of the TIR domain of human TLR1 (crystallised with Zn2+ ions) | | Descriptor: | Toll-like receptor 1 | | Authors: | Vakhrameev, D.D, Luginina, A.P, Shevtsov, M.B, Lushpa, V.A, Mineev, K.S, Borshchevskiy, V.I. | | Deposit date: | 2021-03-15 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of Toll-like receptor 1 intracellular domain structure and activity by Zn 2+ ions.
Commun Biol, 4, 2021
|
|
4GYC
 
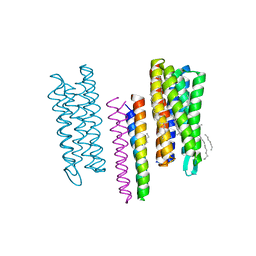 | | Structure of the SRII(D75N mutant)/HtrII Complex in I212121 space group ("U" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2012-09-05 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0501 Å) | | Cite: | Ground state structure of D75N mutant of sensory rhodopsin II in complex with its cognate transducer.
J Photochem Photobiol B, 123C, 2013
|
|
6EYU
 
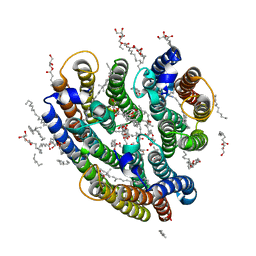 | | Crystal structure of the inward H(+) pump xenorhodopsin | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Shevchenko, V, Polovinkin, V, Mager, T, Gushchin, I, Melnikov, I, Borshchevskiy, V, Popov, A, Alekseev, A, Gordeliy, V. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inward H(+) pump xenorhodopsin: Mechanism and alternative optogenetic approach.
Sci Adv, 3, 2017
|
|
7AV6
 
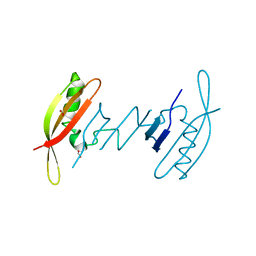 | | FAST in a domain-swapped dimer form | | Descriptor: | FORMIC ACID, Photoactive yellow protein | | Authors: | Bukhdruker, S, Remeeva, A, Ruchkin, D, Gorbachev, D, Povarova, N, Mineev, K, Goncharuk, S, Baranov, M, Mishin, A, Borshchevskiy, V. | | Deposit date: | 2020-11-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NanoFAST: structure-based design of a small fluorogen-activating protein with only 98 amino acids.
Chem Sci, 12, 2021
|
|
6EIG
 
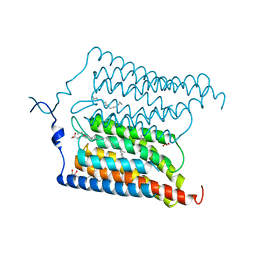 | | Crystal structure of N24Q/C128T mutant of Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, EICOSANE, ... | | Authors: | Kovalev, K, Borshchevskiy, V, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
