3FI1
 
 | | NhaA dimer model | | Descriptor: | Na(+)/H(+) antiporter nhaA | | Authors: | Appel, M, Hizlan, D, Vinothkumar, K.R, Ziegler, C, Kuehlbrandt, W. | | Deposit date: | 2008-12-10 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Conformations of NhaA, the Na/H exchanger from Escherichia coli, in the pH-activated and ion-translocating states
J.Mol.Biol., 386, 2009
|
|
5JOC
 
 | | Crystal structure of the S61A mutant of AmpC BER | | Descriptor: | Beta-lactamase, CITRIC ACID | | Authors: | Na, J.H, An, Y.J, Cha, S.S. | | Deposit date: | 2016-05-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5JXE
 
 | | Human PD-1 ectodomain complexed with Pembrolizumab Fab | | Descriptor: | Pembrolizumab Fab heavy chain, Pembrolizumab Fab light chain, Programmed cell death protein 1 | | Authors: | Na, Z, Bharath, S.R, Song, H. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for blocking PD-1-mediated immune suppression by therapeutic antibody pembrolizumab.
Cell Res., 27, 2017
|
|
5WU7
 
 | |
1SFQ
 
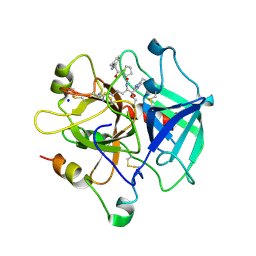 | | Fast form of thrombin mutant R(77a)A bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-20 | | Release date: | 2004-06-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1SG8
 
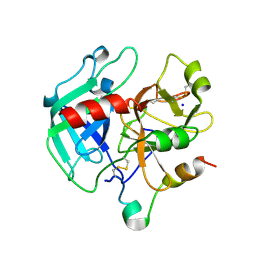 | | Crystal structure of the procoagulant fast form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
2E30
 
 | |
4US4
 
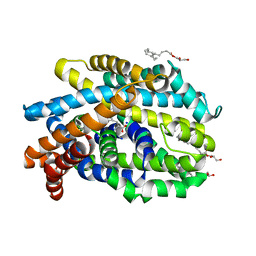 | | Crystal Structure of the Bacterial NSS Member MhsT in an Occluded Inward-Facing State (lipidic cubic phase form) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, SODIUM ION, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
7RPK
 
 | | Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog | | Descriptor: | (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7QTV
 
 | | Beryllium fluoride form of the Na+,K+-ATPase (E2-BeFx) | | Descriptor: | 1-O-decanoyl-beta-D-tagatofuranosyl beta-D-allopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shahsavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
7RPH
 
 | | Cryo-EM structure of murine Dispatched 'R' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPI
 
 | | Cryo-EM structure of murine Dispatched 'T' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
4NYP
 
 | |
4F4R
 
 | | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop
to be published
|
|
4NUK
 
 | | Crystal structure of nickel-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, NICKEL (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUI
 
 | | Crystal structure of cobalt-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, COBALT (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
2FIR
 
 | | Crystal structure of DFPR-VIIa/sTF | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII Heavy Chain (EC 3.4.21.21), ... | | Authors: | Bajaj, S.P, Schmidt, A.E, Padmanabhan, K, Bajaj, M.S, Prevost, D, Schreuder, H. | | Deposit date: | 2005-12-30 | | Release date: | 2006-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Structures of p-Aminobenzamidine- and Benzamidine-VIIa/Soluble Tissue Factor: Unpredicted conformation of the 192-193 peptide bond and mapping of Ca2+, Mg2+, Na+ and Zn2+ sites in factor VIIa
J.Biol.Chem., 281, 2006
|
|
4UET
 
 | | Diversity in the structures and ligand binding sites among the fatty acid and retinol binding proteins of nematodes revealed by Na-FAR-1 from Necator americanus | | Descriptor: | NEMATODE FATTY ACID RETINOID BINDING PROTEIN | | Authors: | Rey-Burusco, M.F, Ibanez Shimabukuro, M, Griffiths, K, Cooper, A, Kennedy, M.W, Corsico, B, Smith, B.O, Griffiths, K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Diversity in the Structures and Ligand Binding Sites of Nematode Fatty Acid and Retinol Binding Proteins Revealed by Na-Far-1 from Necator Americanus.
Biochem.J., 471, 2015
|
|
8V9H
 
 | | GES-5-NA-1-157 complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restricted Rotational Flexibility of the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the GES-5 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
4U9S
 
 | | Crystal structure of NqrC from Vibrio cholerae | | Descriptor: | BROMIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Fritz, G. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
4U9O
 
 | | Crystal structure of NqrA from Vibrio cholerae | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Na(+)-translocating NADH-quinone reductase subunit A | | Authors: | Fritz, G. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
4UAJ
 
 | | Crystal structure of NqrF in hexagonal space group | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, SULFATE ION | | Authors: | Fritz, G. | | Deposit date: | 2014-08-10 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7019 Å) | | Cite: | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
4US3
 
 | | Crystal Structure of the bacterial NSS member MhsT in an Occluded Inward-Facing State | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, SODIUM ION, TRANSPORTER, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
8A1X
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor DQA | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1Y
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
