2FMJ
 
 | |
4IB0
 
 | |
2KLT
 
 | | Second Ca2+ binding domain of NCX1.3 | | Descriptor: | Sodium/calcium exchanger 1 | | Authors: | Hilge, M, Aelen, J, Foarce, A, Perrakis, A, Vuister, G.W. | | Deposit date: | 2009-07-08 | | Release date: | 2009-08-18 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Ca2+ regulation in the Na+/Ca2+ exchanger features a dual electrostatic switch mechanism.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KLS
 
 | | Apo-form of the second Ca2+ binding domain of NCX1.4 | | Descriptor: | Sodium/calcium exchanger 1 | | Authors: | Hilge, M, Aelen, J, Foarce, A, Perrakis, A, Vuister, G.W. | | Deposit date: | 2009-07-08 | | Release date: | 2009-08-18 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Ca2+ regulation in the Na+/Ca2+ exchanger features a dual electrostatic switch mechanism.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7DTC
 
 | | voltage-gated sodium channel Nav1.5-E1784K | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Pan, X, Li, Z. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na v 1.5 reveals the fast inactivation-related segments as a mutational hotspot for the long QT syndrome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2MDF
 
 | |
3X3B
 
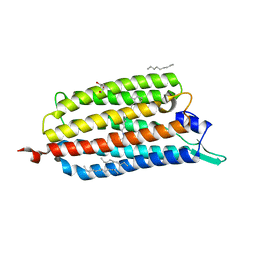 | | Crystal structure of the light-driven sodium pump KR2 in acidic state | | Descriptor: | DI(HYDROXYETHYL)ETHER, OLEIC ACID, RETINAL, ... | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
3X3C
 
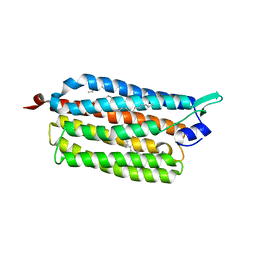 | | Crystal structure of the light-driven sodium pump KR2 in neutral state | | Descriptor: | OLEIC ACID, RETINAL, Sodium pumping rhodopsin | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
5MV2
 
 | | Crystal structure of the E protein of the Japanese encephalitis live attenuated vaccine virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
8H0H
 
 | |
8H2D
 
 | |
3E89
 
 | |
3E8G
 
 | |
4ZPX
 
 | |
6V6T
 
 | |
5F1F
 
 | | Crystal structure of CMY-10 adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, CADMIUM ION | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
7WFW
 
 | | Apo human Nav1.8 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-27 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WEL
 
 | | Human Nav1.8 with A-803467, class II | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WE4
 
 | | Human Nav1.8 with A-803467, class I | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WFR
 
 | | Human Nav1.8 with A-803467, class III | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-27 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2NX9
 
 | |
7YS5
 
 | | RET G-quadruplex in 10mM Na+ | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Yin, S, Cao, C. | | Deposit date: | 2022-08-11 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | SOLUTION NMR | | Cite: | RET G-quadruplex in 10mM Na+
To Be Published
|
|
1HXF
 
 | |
1HXE
 
 | | SERINE PROTEASE | | Descriptor: | HIRUDIN VARIANT-1, RUBIDIUM ION, THROMBIN | | Authors: | Tulinsky, A, Zhang, E. | | Deposit date: | 1995-12-07 | | Release date: | 1996-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular environment of the Na+ binding site of thrombin.
Biophys.Chem., 63, 1997
|
|
7CJ2
 
 | | Crystal structure of the Fab antibody complexed with human YKL-40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 3-like 1 (Cartilage glycoprotein-39), isoform CRA_a, ... | | Authors: | Choi, S, Na, J.H, Lee, S.J, Woo, J.R, Kim, D.Y, Hong, J.T, Lee, W.K. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Fab antibody complexed with human YKL-40
To Be Published
|
|
