3VAH
 
 | | Structure of U2AF65 variant with BrU3C4 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(P*UP*UP*(BRU)P*CP*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAG
 
 | | Structure of U2AF65 variant with BrU3C2 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*U*CP*(BRU)P*UP*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAF
 
 | | Structure of U2AF65 variant with BrU3 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*(BRU)P*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3V4M
 
 | |
3UWT
 
 | |
3ULH
 
 | |
3UD4
 
 | | The C92U mutant c-di-GMP-I riboswitch bound to GpA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*A)-3'), RNA (92-MER), ... | | Authors: | Smith, K.D, Strobel, S.A. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer.
Biochemistry, 51, 2012
|
|
3UD3
 
 | | The C92U mutant c-di-GMP-I riboswitch bound to pGpA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*GP*A)-3'), RNA (92-MER), ... | | Authors: | Smith, K.D, Strobel, S.A. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer.
Biochemistry, 51, 2012
|
|
3UCZ
 
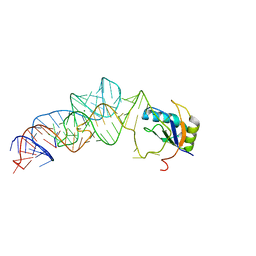 | | The c-di-GMP-I riboswitch bound to GpG | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*G)-3'), RNA (92-MER), ... | | Authors: | Smith, K.D, Strobel, S.A. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer.
Biochemistry, 51, 2012
|
|
3UCU
 
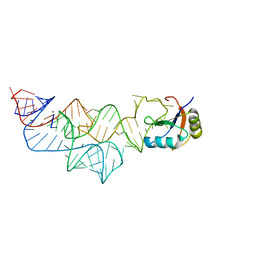 | | The c-di-GMP-I riboswitch bound to pGpG | | Descriptor: | MAGNESIUM ION, RNA (92-MER), U1 small nuclear ribonucleoprotein A, ... | | Authors: | Smith, K.D, Strobel, S.A. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer.
Biochemistry, 51, 2012
|
|
3UCG
 
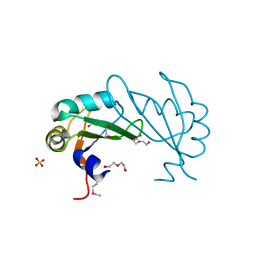 | |
3U1M
 
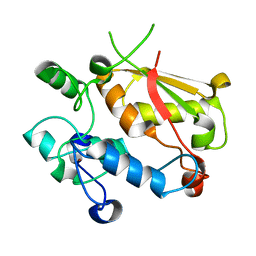 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
3U1L
 
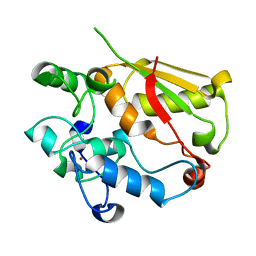 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
3TP2
 
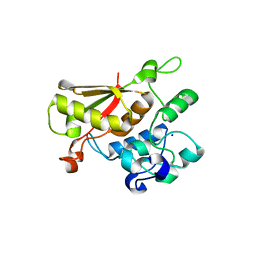 | | Crystal Structure of the Splicing Factor Cwc2 from yeast | | Descriptor: | Pre-mRNA-splicing factor CWC2, SODIUM ION, ZINC ION | | Authors: | Schmitzova, J. | | Deposit date: | 2011-09-07 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein.
Embo J., 31, 2012
|
|
3THT
 
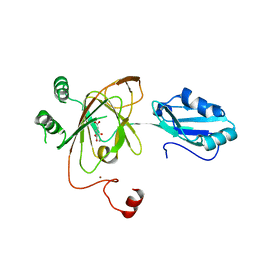 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3THP
 
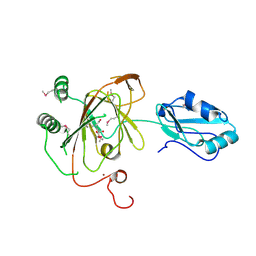 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3SXL
 
 | |
3SMZ
 
 | |
3SDE
 
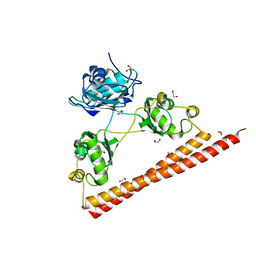 | | Crystal structure of a paraspeckle-protein heterodimer, PSPC1/NONO | | Descriptor: | 1,2-ETHANEDIOL, Non-POU domain-containing octamer-binding protein, Paraspeckle component 1 | | Authors: | Passon, D.M, Lee, M, Bond, C.S. | | Deposit date: | 2011-06-09 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3S8S
 
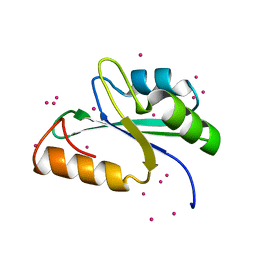 | | Crystal structure of the RRM domain of human SETD1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, UNKNOWN ATOM OR ION | | Authors: | Chao, X, Tempel, W, Bian, C, Cerovina, T, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-30 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the RRM domain of human SETD1A
to be published
|
|
3S7R
 
 | |
3R27
 
 | | Crystal structure of the first RRM domain of heterogeneous nuclear ribonucleoprotein L (HnRNP L) | | Descriptor: | GLYCEROL, Heterogeneous nuclear ribonucleoprotein L | | Authors: | Zhang, W, Liu, Y, Zeng, F, Niu, L, Teng, M, Li, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the first RRM domain of heterogeneous nuclear ribonucleoprotein L (HnRNP L)
To be Published
|
|
3R1L
 
 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Mg2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', Class I ligase ribozyme, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3R1H
 
 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Ca2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', CALCIUM ION, Class I ligase ribozyme, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3Q2T
 
 | | Crystal Structure of CFIm68 RRM/CFIm25/RNA complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6, RNA | | Authors: | Yang, Q, Coseno, M, Gilmartin, G.M, Doublie, S. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.061 Å) | | Cite: | Crystal Structure of a Human Cleavage Factor CFI(m)25/CFI(m)68/RNA Complex Provides an Insight into Poly(A) Site Recognition and RNA Looping.
Structure, 19, 2011
|
|
