1B6T
 
 | |
1B5S
 
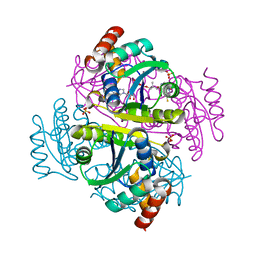
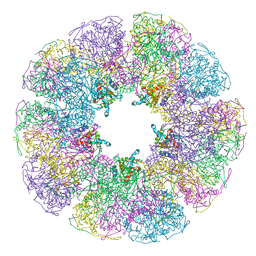 | | DIHYDROLIPOYL TRANSACETYLASE (E.C.2.3.1.12) CATALYTIC DOMAIN (RESIDUES 184-425) FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Izard, T, Aevarsson, A, Allen, M.D, Westphal, A.H, Perham, R.N, De Kok, A, Hol, W.G. | | Deposit date: | 1999-01-10 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6U4K
 
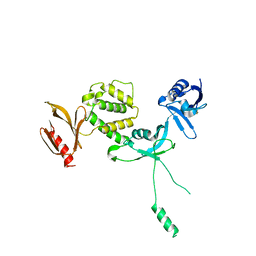 | | Human talin2 residues 1-403 | | Descriptor: | Talin-2 | | Authors: | Izard, T, Rangarajan, E.S, Colgan, L, Yasuda, R, Chinthalapudi, K. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (2.555 Å) | | Cite: | A distinct talin2 structure directs isoform specificity in cell adhesion.
J.Biol.Chem., 295, 2020
|
|
4EHP
 
 | |
4IGG
 
 | |
6MFS
 
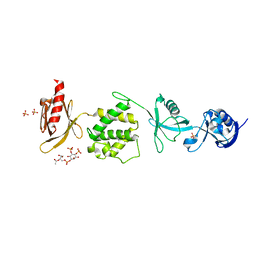 | | Mouse talin1 residues 1-138 fused to residues 169-400 in complex with phosphatidylinositol 4,5-bisphosphate (PIP2) | | Descriptor: | PHOSPHATE ION, Talin-1 fusion, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Izard, T, Chinthalapudi, K, Rangarajan, E.S. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3MYI
 
 | |
2GWW
 
 | |
2HSQ
 
 | |
2IBF
 
 | |
1GN8
 
 | |
1H1T
 
 | |
1SYQ
 
 | |
1YDI
 
 | |
7KTU
 
 | |
7KTV
 
 | |
7KTT
 
 | |
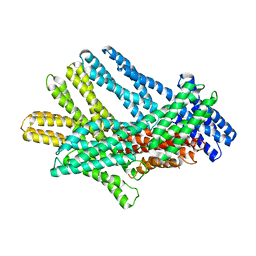
7KTW
 
 | |
1NAL
 
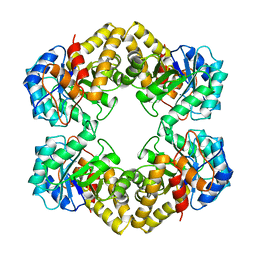 | | THE THREE-DIMENSIONAL STRUCTURE OF N-ACETYLNEURAMINATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | N-ACETYLNEURAMINATE LYASE, SULFATE ION | | Authors: | Izard, T, Lawrence, M.C, Malby, R.L, Lilley, G.G, Colman, P.M. | | Deposit date: | 1994-02-28 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure of N-acetylneuraminate lyase from Escherichia coli.
Structure, 2, 1994
|
|
1QHX
 
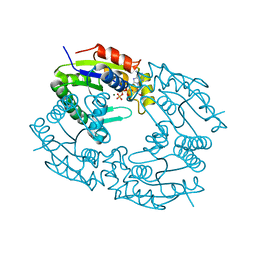 | |
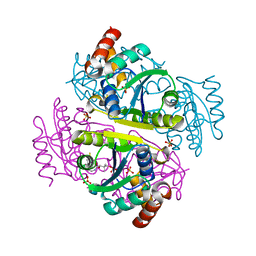
1QJC
 
 | |
1QHN
 
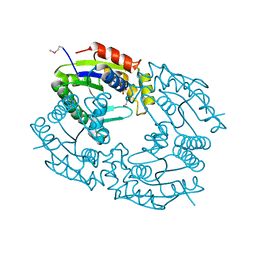 | | CHLORAMPHENICOL PHOSPHOTRANSFERASE FROM STREPTOMYCES VENEZUELAE | | Descriptor: | CHLORAMPHENICOL PHOSPHOTRANSFERASE, SULFATE ION | | Authors: | Izard, T. | | Deposit date: | 1999-05-23 | | Release date: | 2000-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structures of Chloramphenicol Phosphotransferase Reveal a Novel Inactivation Mechanism
Embo J., 19, 2000
|
|
1QHS
 
 | |
1QHY
 
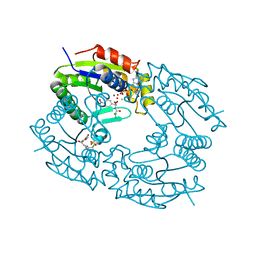 | |
1RKC
 
 | | Human vinculin head (1-258) in complex with talin's vinculin binding site 3 (residues 1944-1969) | | Descriptor: | Talin, Vinculin | | Authors: | Izard, T, Evans, G, Borgon, R.A, Rush, C.L, Bricogne, G, Bois, P.R. | | Deposit date: | 2003-11-21 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Vinculin activation by talin through helical bundle conversion
Nature, 427, 2004
|
|
