1NFD
 
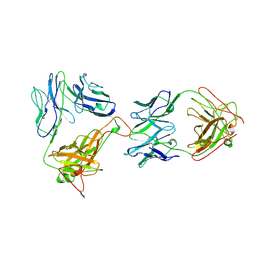 | | AN ALPHA-BETA T CELL RECEPTOR (TCR) HETERODIMER IN COMPLEX WITH AN ANTI-TCR FAB FRAGMENT DERIVED FROM A MITOGENIC ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H57 FAB, N15 ALPHA-BETA T-CELL RECEPTOR | | Authors: | Wang, J.-H, Lim, K, Smolyar, A, Teng, M.-K, Sacchittini, J, Reinherz, E.L. | | Deposit date: | 1997-08-04 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody.
EMBO J., 17, 1998
|
|
3LIQ
 
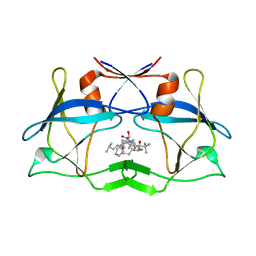 | | Crystal Structure of HTLV protease complexed with the inhibitor, KNI-10673 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-5,5-dimethyl-N-(2-methylpropyl)-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LIX
 
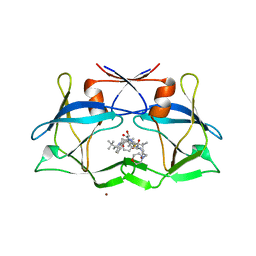 | | crystal structure of htlv protease complexed with the inhibitor KNI-10729 | | Descriptor: | N-{(1S,2S)-1-benzyl-3-[(4R)-5,5-dimethyl-4-{[(1R)-1,2,2-trimethylpropyl]carbamoyl}-1,3-thiazolidin-3-yl]-2-hydroxy-3-oxopropyl}-3-methyl-N~2~-{(2S)-2-[(morpholin-4-ylacetyl)amino]-2-phenylacetyl}-L-valinamide, Protease, ZINC ION | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LIV
 
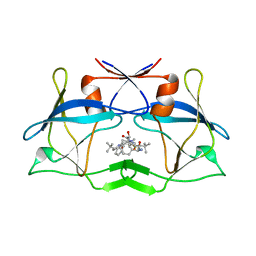 | | crystal structure of HTLV protease complexed with the inhibitor KNI-10683 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-N-[(2R)-3,3-dimethylbutan-2-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LIT
 
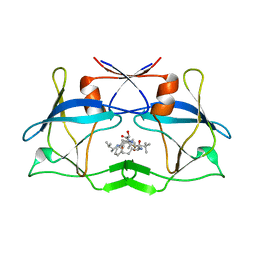 | | The crystal structure of htlv protease complexed with the inhibitor KNI-10681 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-5,5-dimethyl-N-[(2R)-3-methylbutan-2-yl]-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
1S97
 
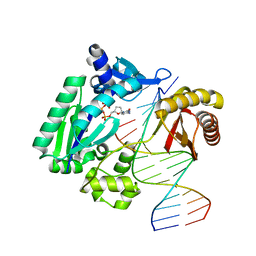 | | DPO4 with GT mismatch | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*G)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
1Q1T
 
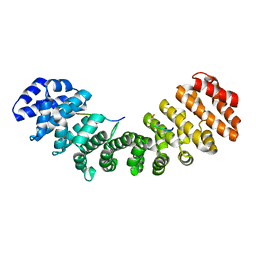 | | Mouse Importin alpha: non-phosphorylated SV40 CN peptide complex | | Descriptor: | Importin alpha-2 subunit, Large T antigen | | Authors: | Fontes, M.R.M, Teh, T, Toth, G, John, A, Pavo, I, Jans, D.A, Kobe, B. | | Deposit date: | 2003-07-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha
Biochem.J., 375, 2003
|
|
1SY6
 
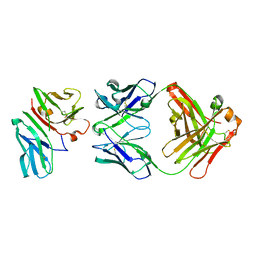 | | Crystal Structure of CD3gammaepsilon Heterodimer in Complex with OKT3 Fab Fragment | | Descriptor: | OKT3 Fab heavy chain, OKT3 Fab light chain, T-cell surface glycoprotein CD3 gamma/epsilon chain | | Authors: | Kjer-Nielsen, L, Dunstone, M.A, Kostenko, L, Ely, L.K, Beddoe, T, Misfud, N.A, Purcell, A.W, Brooks, A.G, McCluskey, J, Rossjohn, J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-05-25 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the human T cell receptor CD3(epsilon)(gamma) heterodimer complexed to the therapeutic mAb OKT3.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4DZB
 
 | |
1Q1S
 
 | | Mouse Importin alpha- phosphorylated SV40 CN peptide complex | | Descriptor: | Importin alpha-2 subunit, Large T antigen | | Authors: | Fontes, M.R.M, Teh, T, Toth, G, John, A, Pavo, I, Jans, D.A, Kobe, B. | | Deposit date: | 2003-07-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha
Biochem.J., 375, 2003
|
|
1S9F
 
 | | DPO with AT matched | | Descriptor: | 2',3'-DIDEOXYCYTOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-04 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
2J3Q
 
 | | Torpedo acetylcholinesterase complexed with fluorophore thioflavin T | | Descriptor: | 2-[4-(DIMETHYLAMINO)PHENYL]-6-HYDROXY-3-METHYL-1,3-BENZOTHIAZOL-3-IUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Harel, M, Cusack, B, Johnson, J.L, Silman, I, Sussman, J.L, Rosenberry, T.L. | | Deposit date: | 2006-08-23 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of thioflavin T bound to the peripheral site of Torpedo californica acetylcholinesterase reveals how thioflavin T acts as a sensitive fluorescent reporter of ligand binding to the acylation site.
J. Am. Chem. Soc., 130, 2008
|
|
2IS3
 
 | |
2KHY
 
 | |
4EI6
 
 | | Structure of XV19 Valpha1-Vbeta16 Type-II Natural Killer T cell receptor | | Descriptor: | Valpha1 XV19 Type II Natural Killer T cell receptor (mouse variable domain, human constant domain), Vbeta16 XV19 Type II Natural Killer T cell receptor (mouse variable domain | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor.
Nat.Immunol., 13, 2012
|
|
3V9U
 
 | |
3VA3
 
 | |
3V9X
 
 | |
2C7U
 
 | | Conflicting selective forces affect CD8 T-cell receptor contact sites in an HLA-A2 immunodominant HIV epitope. | | Descriptor: | BETA-2-MICROGLOBULIN, GAG PROTEIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Iversen, A.K, Stewart-Jones, G, Learn, G.H, Christie, N, Sylvester-Hviid, C, Armitage, A.E, Kaul, R, Beattie, T, Lee, J.K, Li, Y, Chotiyarnwong, P, Dong, T, Xu, X, Luscher, M.A, MacDonald, K, Ullum, H, Klarlund-Pedersen, B, Skinhoj, P, Fugger, J.L, Buus, S, Mullins, J.I, Jones, E.Y, van der Merwe, P.A, McMichael, A.J. | | Deposit date: | 2005-11-29 | | Release date: | 2006-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Conflicting Selective Forces Affect T Cell Receptor Contacts in an Immunodominant Human Immunodeficiency Virus Epitope.
Nat.Immunol., 7, 2006
|
|
3V9W
 
 | |
3B0W
 
 | | Crystal structure of the orphan nuclear receptor ROR(gamma)t ligand-binding domain in complex with digoxin | | Descriptor: | DIGOXIN, Nuclear receptor ROR-gamma | | Authors: | Fujita-Sato, S, Ito, S, Isobe, T, Ohyama, T, Wakabayashi, K, Morishita, K, Ando, O, Isono, F. | | Deposit date: | 2011-06-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Digoxin That Antagonizes ROR{gamma}t Receptor Activity and Suppresses Th17 Cell Differentiation and Interleukin (IL)-17 Production
J.Biol.Chem., 286, 2011
|
|
2YRP
 
 | | Solution structure of the TIG domain from Human Nuclear factor of activated T-cells, cytoplasmic 4 | | Descriptor: | Nuclear factor of activated T-cells, cytoplasmic 4 | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TIG domain from Human Nuclear factor of activated T-cells, cytoplasmic 4
To be Published
|
|
3V9Z
 
 | |
3V9S
 
 | |
3VA0
 
 | |
