6P5W
 
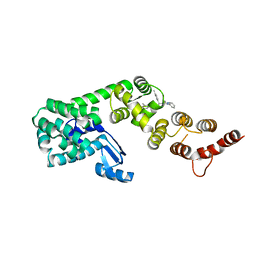 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6P5V
 
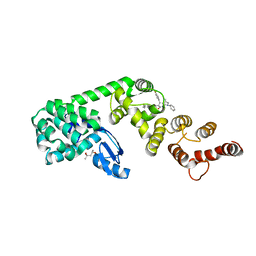 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
7AUX
 
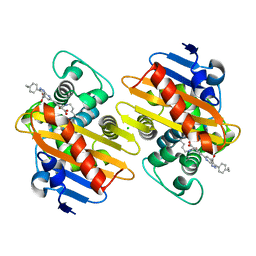 | | Crystal structure of OXA-48 beta-lactamase in the complex with the inhbitor ID2 | | Descriptor: | 6-(4-carboxyphenyl)-3-(4-ethylphenyl)-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Beta-lactamase, CHLORIDE ION | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Garofalo, B, Ombrato, R. | | Deposit date: | 2020-11-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Novel Chemical Series of OXA-48 beta-Lactamase Inhibitors by High-Throughput Screening.
Pharmaceuticals, 14, 2021
|
|
6GRQ
 
 | |
8R5K
 
 | |
6CT2
 
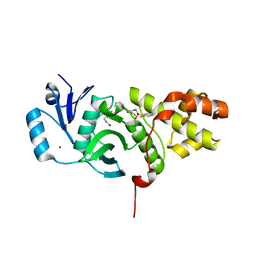 | | MYST histone acetyltransferase KAT6A/B in complex with WM-1119 | | Descriptor: | 3-fluoro-N'-[(2-fluorophenyl)sulfonyl]-5-(pyridin-2-yl)benzohydrazide, Histone acetyltransferase KAT8, MAGNESIUM ION, ... | | Authors: | Ren, B, Peat, T.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
7YST
 
 | |
6S2Y
 
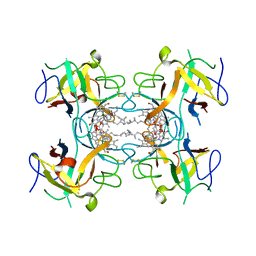 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-soluble chlorophyll protein | | Authors: | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | Deposit date: | 2019-06-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
4GJ0
 
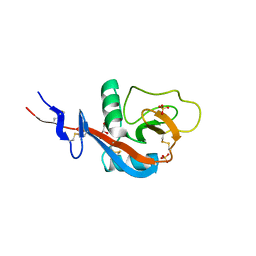 | | Crystal structure of CD23 lectin domain mutant S252A | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor, SULFATE ION | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-09 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
4GK1
 
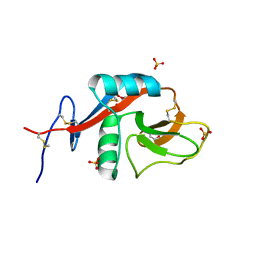 | | Crystal structure of CD23 lectin domain mutant D270A | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor, SULFATE ION | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-10 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
6M95
 
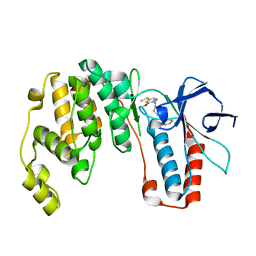 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping: compound 1 | | Descriptor: | (4-benzylpiperidin-1-yl)[2-methoxy-4-(methylsulfanyl)phenyl]methanone, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
8GCI
 
 | |
8AAE
 
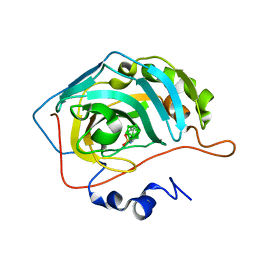 | | CAII in complex with para-carboran-propylsulfonamid | | Descriptor: | Carbonic anhydrase 2, Para-Carborane propyl-sulfonamide, ZINC ION | | Authors: | Brynda, J, Rezacova, P. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | B-H⋯ pi and C-H⋯ pi interactions in protein-ligand complexes: carbonic anhydrase II inhibition by carborane sulfonamides.
Phys Chem Chem Phys, 25, 2023
|
|
8AA6
 
 | | CAII in complex with meta-carboran-propylsulfonamid | | Descriptor: | Carbonic anhydrase 2, ZINC ION, meta-carboran-propylsulfonamid | | Authors: | Brynda, J, Rezacova, P, Kugler, M. | | Deposit date: | 2022-06-30 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | B-H⋯ pi and C-H⋯ pi interactions in protein-ligand complexes: carbonic anhydrase II inhibition by carborane sulfonamides.
Phys Chem Chem Phys, 25, 2023
|
|
5Z0S
 
 | | Crystal structure of FGFR1 kinase domain in complex with a novel inhibitor | | Descriptor: | 1-[(6-chloroimidazo[1,2-b]pyridazin-3-yl)sulfonyl]-6-(1-methyl-1H-pyrazol-4-yl)-1H-pyrazolo[4,3-b]pyridine, Fibroblast growth factor receptor 1 | | Authors: | Liu, Q, Xu, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Discovery of a Series of 5H-Pyrrolo[2,3-b]pyrazine FGFR Kinase Inhibitors
Molecules, 23, 2018
|
|
6TH4
 
 | | Tubulin-inhibitor complex | | Descriptor: | 1,2,3,9-tetramethoxy-6-methylidene-5~{H}-cyclohepta[a]naphthalen-8-one, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Varela, P.F, Gigant, B. | | Deposit date: | 2019-11-18 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | B-nor-methylene Colchicinoid PT-100 Selectively Induces Apoptosis in Multidrug-Resistant Human Cancer Cells via an Intrinsic Pathway in a Caspase-Independent Manner
Acs Omega, 7, 2022
|
|
6DQB
 
 | | LINKED KDM5A JMJ DOMAIN FORMING COVALENT BOND TO INHIBITOR N71 i.e. 2-((3-(4-(dimethylamino)but-2-enamido)phenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 2-{(R)-(3-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(R)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(S)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6RX4
 
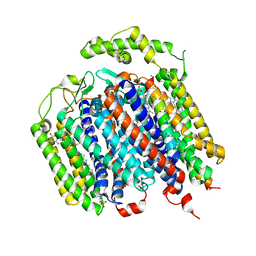 | | THE STRUCTURE OF BD OXIDASE FROM ESCHERICHIA COLI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-I ubiquinol oxidase subunit 1, ... | | Authors: | Rasmussen, T, Boettcher, B, Thesseling, A, Friedrich, T. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Homologous bd oxidases share the same architecture but differ in mechanism.
Nat Commun, 10, 2019
|
|
5MNS
 
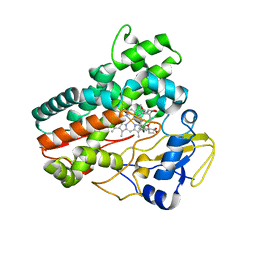 | | Structural and functional characterization of OleP in complex with 6DEB in sodium formate | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Parisi, G, Savino, C, Montemiglio, L.C, Vallone, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Substrate-induced conformational change in cytochrome P450 OleP.
FASEB J., 33, 2019
|
|
6S2Z
 
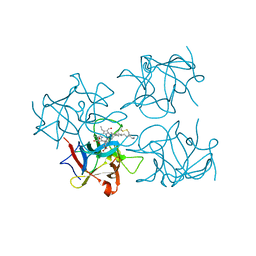 | | Water-soluble Chlorophyll Protein (WSCP) from Brassica oleracea var. Botrytis with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-Soluble Chlorophyll Protein | | Authors: | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | Deposit date: | 2019-06-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
5GV5
 
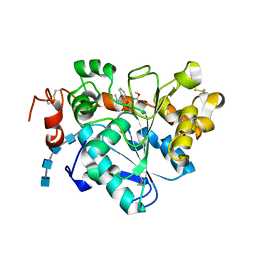 | | Crystal structure of Candida antarctica Lipase B with active Ser105 modified with a phosphonate inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, ... | | Authors: | Park, S.Y, Lee, H. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural and Experimental Evidence for the Enantiomeric Recognition toward a Bulky sec-Alcohol by Candida antarctica Lipase B
Acs Catalysis, 6, 2016
|
|
6VJT
 
 | |
8OKV
 
 | | lipoprotein BT2095 from Bacteroides thetaiotamicron bound to cyanocobalamin CnCbl | | Descriptor: | CYANOCOBALAMIN, Putative surface layer protein | | Authors: | Javier Abellon-Ruiz, J, van den Berg, B, Jana, K, Kleinekathofer, U, Silale, A, Frey, A.M, Basle, A, Trost, M. | | Deposit date: | 2023-03-29 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
7O25
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE from T. maritima (reaction triggered in the crystal) | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O26
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (5'dA + Methionine) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
