5CZF
 
 | |
6YAU
 
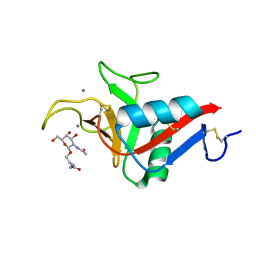 | | CRYSTAL STRUCTURE OF ASGPR 1 IN COMPLEX WITH GN-A. | | Descriptor: | 5-[(2~{R},3~{R},4~{R},5~{R},6~{R})-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-~{N}-[3-(propanoylamino)propyl]pentanamide, Asialoglycoprotein receptor 1, CALCIUM ION | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2020-03-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | Triantennary GalNAc Molecular Imaging Probes for Monitoring Hepatocyte Function in a Rat Model of Nonalcoholic Steatohepatitis.
Adv Sci, 7, 2020
|
|
6YNZ
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo composite tetramer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
1XYD
 
 | | NMR Solution Structure of Rat Zinc-Calcium-S100B, 20 Structures | | Descriptor: | CALCIUM ION, S-100 protein, beta chain, ... | | Authors: | Wilder, P.T, Varney, K.M, Weber, D.J. | | Deposit date: | 2004-11-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Zinc- and Calcium-Bound Rat S100B as Determined by Nuclear Magnetic Resonance Spectroscopy
Biochemistry, 44, 2005
|
|
5XJG
 
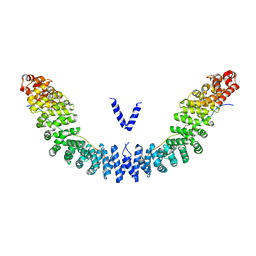 | | Crystal structure of Vac8p bound to Nvj1p | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Nucleus-vacuole junction protein 1, ... | | Authors: | Jeong, H, Park, J, Jun, Y, Lee, C. | | Deposit date: | 2017-05-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YC5
 
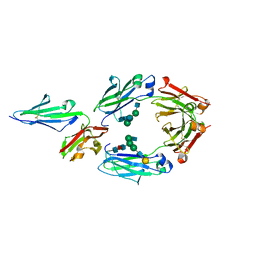 | | Crystal structure of human IgG-Fc in complex with aglycan and optimized Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Tsumoto, K, Kiyoshi, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Assessing the Heterogeneity of the Fc-Glycan of a Therapeutic Antibody Using an engineered Fc gamma Receptor IIIa-Immobilized Column.
Sci Rep, 8, 2018
|
|
8SAS
 
 | | CryoEM structure of DH270.5-CH848.10.17 | | Descriptor: | CH848.10.17 gp120, CH848.10.17 gp41, DH270.5 variable heavy chian, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8TUX
 
 | |
1AGO
 
 | |
3O3F
 
 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and magnesium ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MAGNESIUM ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
3O3H
 
 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and manganese ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MANGANESE (II) ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
3O3G
 
 | | T. maritima RNase H2 in complex with nucleic acid substrate and calcium ions | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
8CO1
 
 | | Type II Secretion System | | Descriptor: | IPT/TIG domain-containing protein, Lipoprotein, Probable type IV piliation system protein DR_0774 | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2023-02-26 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural characterization and functional insights into the type II secretion system of the poly-extremophile Deinococcus radiodurans.
J.Biol.Chem., 300, 2024
|
|
6OZC
 
 | | BG505 SOSIP.664 with 2G12 Fab2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 Fab Heavy chain, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions.
Structure, 28, 2020
|
|
8DJ7
 
 | | The complex structure between human IgG1 Fc and its high affinity receptor FcgRI H174R variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin gamma Fc receptor I, Ig gamma-1 Fc chain, ... | | Authors: | Lu, J, Sun, P.D. | | Deposit date: | 2022-06-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fc gamma RI FG-loop functions as a pH sensitive switch for IgG binding and release.
Front Immunol, 14, 2023
|
|
8CZZ
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with Temsavir, 8ANC195, and 10-1074 | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
4WO5
 
 | | Crystal structure of a BRAF kinase domain monomer | | Descriptor: | N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Critton, D.A. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of a BRAF kinase domain monomer explains basis for allosteric regulation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4X4M
 
 | | Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high affinity IgG binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lu, J, Sun, P.D. | | Deposit date: | 2014-12-03 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.485 Å) | | Cite: | Structure of Fc gamma RI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6Q3G
 
 | | Structure of native bacteriophage P68 | | Descriptor: | Arstotzka protein, Head fiber protein, Inner core protein, ... | | Authors: | Dominik, H, Karel, S, Fuzik, T, Plevka, P. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
6Q10
 
 | |
8DVD
 
 | |
8DOK
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
7L56
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-43 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
6BF4
 
 | |
6CH7
 
 | |
