8CO1
 
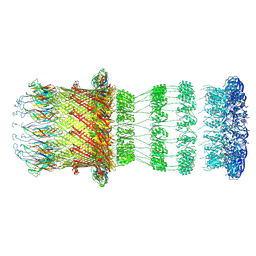 | | Type II Secretion System | | Descriptor: | IPT/TIG domain-containing protein, Lipoprotein, Probable type IV piliation system protein DR_0774 | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2023-02-26 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural characterization and functional insights into the type II secretion system of the poly-extremophile Deinococcus radiodurans.
J.Biol.Chem., 300, 2024
|
|
8ACA
 
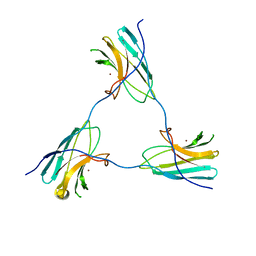 | | SDBC DR_0644 subunit, only-Cu Superoxide Dismutase | | Descriptor: | COPPER (II) ION, DR_0644, only-Cu Superoxide Dismutase | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
8ACQ
 
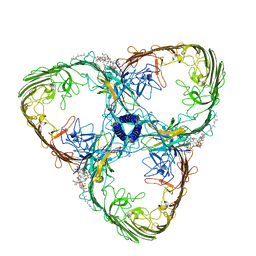 | | S-layer Deinoxanthin-Binding Complex (SDBC), subunit DR_2577 assembled with its SOD DR_0644 | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, DR_0644, ... | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
8AGD
 
 | | Full SDBC and SOD assembly | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, FE (III) ION, ... | | Authors: | Farci, D, Graca, A.T, Piano, D. | | Deposit date: | 2022-07-19 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
7ZGX
 
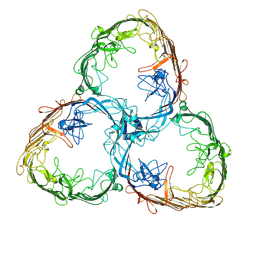 | | S-layer Deinoxanthin Binding Complex, C1 symmetry | | Descriptor: | S-layer protein SlpA | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-04-04 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The cryo-EM structure of the S-layer deinoxanthin-binding complex of Deinococcus radiodurans informs properties of its environmental interactions.
J.Biol.Chem., 298, 2022
|
|
7ZGY
 
 | | S-layer Deinoxanthin Binding Complex, C3 symmetry | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, FE (III) ION, ... | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-04-04 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The cryo-EM structure of the S-layer deinoxanthin-binding complex of Deinococcus radiodurans informs properties of its environmental interactions.
J.Biol.Chem., 298, 2022
|
|
