8E26
 
 | | Crystal Structure of SARS-CoV-2 Main Protease N142S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-08-14 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8EF4
 
 | |
5V5T
 
 | | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Conserved domain protein | | Authors: | De Groote, M.C.R, Camargo, I.L, Serror, P, Horjales, E. | | Deposit date: | 2017-03-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and functional characterization of ElrR, a member of the RRNPP family of transcriptional regulators.
To be published
|
|
8EC6
 
 | | Cryo-EM structure of the Glutaminase C core filament (fGAC) | | Descriptor: | Isoform 2 of Glutaminase kidney isoform, mitochondrial, PHOSPHATE ION | | Authors: | Ambrosio, A.L, Dias, S.M, Quesnay, J.E, Portugal, R.V, Cassago, A, van Heel, M.G, Islam, Z, Rodrigues, C.T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of glutaminase activation through filamentation and the role of filaments in mitophagy protection.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EYJ
 
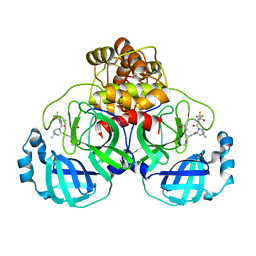 | | Crystal Structure of uncleaved SARS-CoV-2 Main Protease C145S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | An in-solution snapshot of SARS-COV-2 main protease maturation process and inhibition.
Nat Commun, 14, 2023
|
|
5WFI
 
 | |
5V5U
 
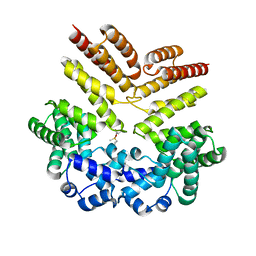 | | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis | | Descriptor: | Conserved domain protein | | Authors: | De Groote, M.C.R, Camargo, I.L, Serror, P, Horjales, E. | | Deposit date: | 2017-03-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis
To be published
|
|
8SCC
 
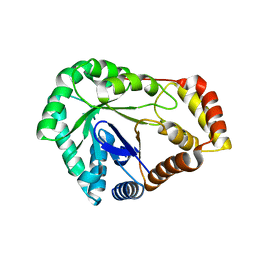 | | Crystal Structure of L-galactose 1-dehydrogenase de Myrciaria dubia | | Descriptor: | L-galactose dehydrogenase | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-05 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
3LMN
 
 | | Oligomeric structure of the DUSP domain of human USP15 | | Descriptor: | ACETIC ACID, FORMIC ACID, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Walker, J.R, Asinas, A, Avvakumov, G.V, Alenkin, D, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2010-01-31 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Ubiquitin-Specific Protease 15 DUSP Domain
To be Published
|
|
8SJJ
 
 | | X-ray structure of the metastable SEPT14-SEPT7 heterodimeric coiled coil | | Descriptor: | DI(HYDROXYETHYL)ETHER, SODIUM ION, Septin 7, ... | | Authors: | Cavini, I.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | X-ray structure of the metastable SEPT14-SEPT7 coiled coil reveals a hendecad region crucial for heterodimerization.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SG0
 
 | | Crystal Structure of GDP-manose 3,5 epimerase de Myrciaria dubia in complex with substrate, product and NAD | | Descriptor: | GDP-mannose 3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-DIPHOSPHATE-BETA-L-GALACTOSE, ... | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
8SKB
 
 | | Crystal Structure of GDP-mannose 3,5 epimerase de Myrciaria dubia in complex with NAD | | Descriptor: | GDP-mannose 3,5 epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-19 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
8UM3
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
8U4A
 
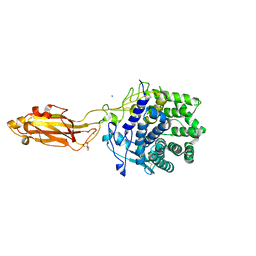 | |
8U4F
 
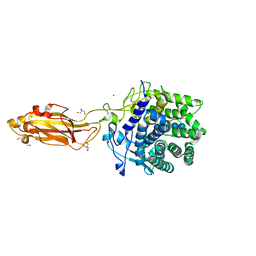 | |
8U49
 
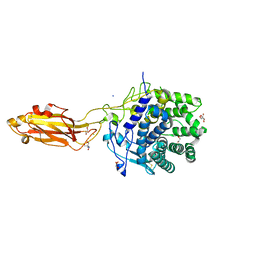 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
8USU
 
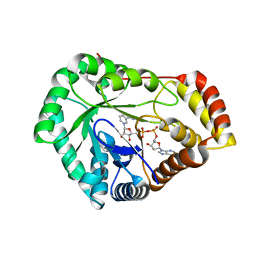 | | Crystal Structure of L-galactose 1-dehydrogenase of Myrciaria dubia in complex with NAD | | Descriptor: | L-galactose dehydrogenase isoform X1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
3OLQ
 
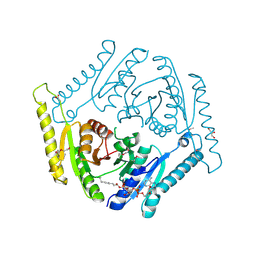 | | The crystal structure of a universal stress protein E from Proteus mirabilis HI4320 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | The crystal structure of a universal stress protein E from
Proteus mirabilis HI4320
To be Published
|
|
8SGD
 
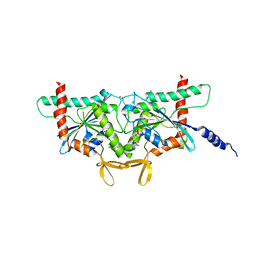 | | Crystal Structure of CDC3(G) - CDC10(Delta 1-10) heterocomplex from Saccharomyces cerevisiae | | Descriptor: | CDC10 isoform 1, CDC3 isoform 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Saladino, G.C.R, Leonardo, D.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A key piece of the puzzle: The central tetramer of the Saccharomyces cerevisiae septin protofilament and its implications for self-assembly.
J.Struct.Biol., 215, 2023
|
|
5LJM
 
 | | Structure of SPATA2 PUB domain | | Descriptor: | GLYCEROL, Spermatogenesis-associated protein 2 | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
5LJN
 
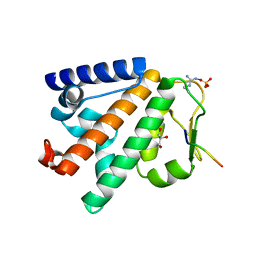 | | Structure of the HOIP PUB domain bound to SPATA2 PIM peptide | | Descriptor: | E3 ubiquitin-protein ligase RNF31, GLYCEROL, SULFATE ION, ... | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
6KCZ
 
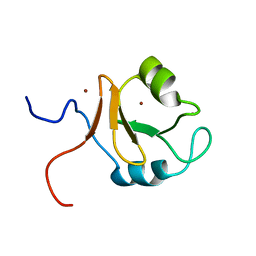 | |
6KHV
 
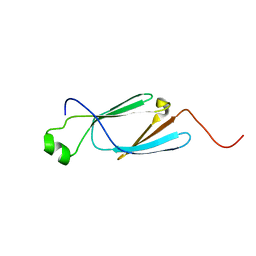 | | Solution Structure of the CS2 Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
6K7W
 
 | | Solution Structure of the CS1 Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
