4ZUU
 
 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-CF9 | | Descriptor: | Beta-2-microglobulin, CYS-THR-SER-GLU-GLU-MET-ASN-ALA-PHE, Classical MHC class I antigen | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
6CHH
 
 | | Structure of human NNMT in complex with bisubstrate inhibitor MS2756 | | Descriptor: | (2~{S})-5-[2-(3-aminocarbonylphenyl)ethyl-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]amino]-2-azanyl-pentanoic acid, 1,2-ETHANEDIOL, Nicotinamide N-methyltransferase | | Authors: | Babault, N, Liu, J, Jin, J. | | Deposit date: | 2018-02-22 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Bisubstrate Inhibitors of Nicotinamide N-Methyltransferase (NNMT).
J. Med. Chem., 61, 2018
|
|
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
7SIN
 
 | | Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor | | Descriptor: | 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, Isoform 1 of Extracellular calcium-sensing receptor | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1NEZ
 
 | | The Crystal Structure of a TL/CD8aa Complex at 2.1A resolution:Implications for Memory T cell Generation, Co-receptor Preference and Affinity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, Y, Xiong, Y, Naidenko, O.V, Liu, J.H, Zhang, R, Joachimiak, A, Kronenberg, M, Cheroutre, H, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2002-12-12 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a TL/CD8alphaalpha Complex at 2.1 A resolution: Implications for modulation of T cell activation and memory
Immunity, 18, 2003
|
|
1D9K
 
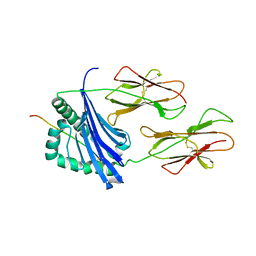 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | Authors: | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | Deposit date: | 1999-10-28 | | Release date: | 1999-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
8K0S
 
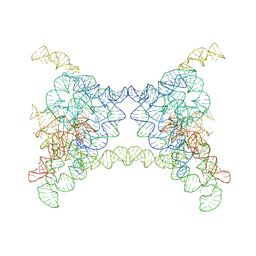 | |
8K0P
 
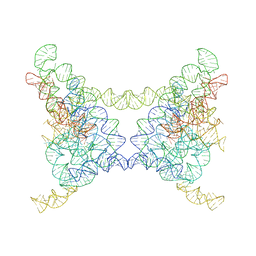 | |
8K0R
 
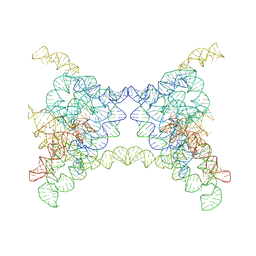 | |
8K15
 
 | |
8K0Q
 
 | |
5MEI
 
 | | Crystal structure of Agelastatin A bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | McClary, B, Zinshteyn, B, Meyer, M, Jouanneau, M, Pellegrino, S, Yusupova, G, Schuller, A, Reyes, J.C.P, Lu, J, Luo, C, Dang, Y, Romo, D, Yusupov, M, Green, R, Liu, J.O. | | Deposit date: | 2016-11-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of Eukaryotic Translation by the Antitumor Natural Product Agelastatin A.
Cell Chem Biol, 24, 2017
|
|
5K0K
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2434 | | Descriptor: | 15-{4-[(4-methylpiperazin-1-yl)methyl]phenyl}-4,5,6,7,9,10,11,12-octahydro-2,16-(azenometheno)pyrrolo[2,1-d][1,3,5,9]te traazacyclotetradecin-8(3H)-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, X, Liu, J, Zhang, W, Stashko, M.A, Nichols, J, DeRyckere, D, Miley, M.J, Norris-Drouin, J, Chen, Z, Machius, M, Wood, E, Graham, D.K, Earp, H.S, Graham, K, Kireev, D, Frye, S.V. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Design and Synthesis of Novel Macrocyclic Mer Tyrosine Kinase Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
6OCR
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OCT
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1M63
 
 | | Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes | | Descriptor: | CALCINEURIN B SUBUNIT ISOFORM 1, CALCIUM ION, CYCLOSPORIN A, ... | | Authors: | Huai, Q, Kim, H.-Y, Liu, Y, Zhao, Y, Mondragon, A, Liu, J.O, Ke, H. | | Deposit date: | 2002-07-12 | | Release date: | 2002-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Calcineurin-Cyclophilin-Cyclosporin Shows Common But Distinct Recognition of Immunophilin-Drug Complexes
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6XDM
 
 | |
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
1BQS
 
 | | THE CRYSTAL STRUCTURE OF MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 (MADCAM-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1) | | Authors: | Tan, K, Casasnovas, J.M, Liu, J.H, Briskin, M.J, Springer, T.A, Wang, J.-H. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition.
Structure, 6, 1998
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
1IC1
 
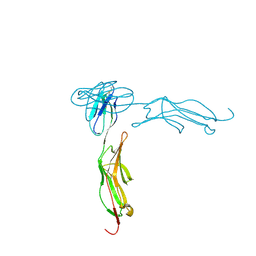 | | THE CRYSTAL STRUCTURE FOR THE N-TERMINAL TWO DOMAINS OF ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-1 | | Authors: | Casasnovas, J.M, Stehle, T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1EI8
 
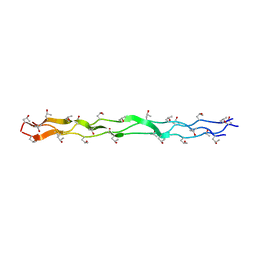 | |
8J5D
 
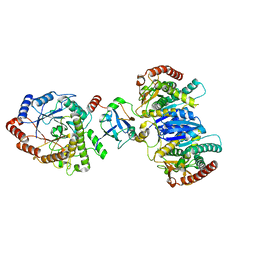 | | Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH | | Descriptor: | Beta-amylase 1, chloroplastic, Malate dehydrogenase, ... | | Authors: | Guan, Z.Y, Liu, J, Yan, J.J. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit beta-amylase to promote starch degradation.
Plant Cell, 36, 2023
|
|
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | Authors: | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
