4U8B
 
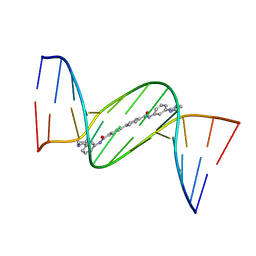 | | Crystal structure of D(CGCGAATTCGCG)2 complexed with BPH-1358 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, N,N'-bis[3-(4,5-dihydro-1H-imidazol-2-yl)phenyl]biphenyl-4,4'-dicarboxamide | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
4U8C
 
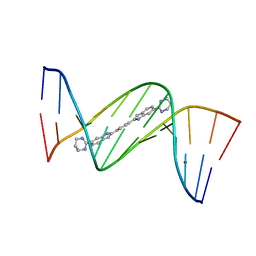 | | Crystal structure of D(CGCGAATTCGCG)2 complexed with BPH-1409 | | Descriptor: | 2,2'-benzene-1,4-diylbis[6-(1,4,5,6-tetrahydropyrimidin-2-yl)-1H-indole], DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
4U82
 
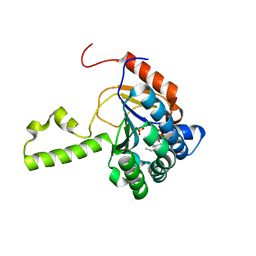 | | Structure of S. aureus undecaprenyl diphosphate synthase in complex with FSPP and sulfate | | Descriptor: | Isoprenyl transferase, MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
4U8A
 
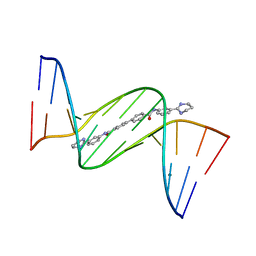 | | Crystal structure of D(CGCGAATTCGCG)2 complexed with BPH-1503 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, N,N'-bis[3-(1,4,5,6-tetrahydropyrimidin-2-yl)phenyl]biphenyl-4,4'-dicarboxamide | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
6GQ3
 
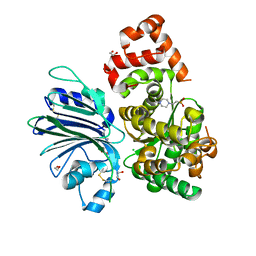 | | Human asparagine synthetase (ASNS) in complex with 6-diazo-5-oxo-L-norleucine (DON) at 1.85 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-OXO-L-NORLEUCINE, ... | | Authors: | Zhu, W, Radadiya, A, Bisson, C, Jin, Y, Nordin, B.E, Imasaki, T, Wenzel, S, Sedelnikova, S.E, Berry, A.H, Nomanbhoy, T.K, Kozarich, J.W, Takagi, Y, Rice, D.W, Richards, N.G.J. | | Deposit date: | 2018-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution crystal structure of human asparagine synthetase enables analysis of inhibitor binding and selectivity.
Commun Biol, 2, 2019
|
|
1PFT
 
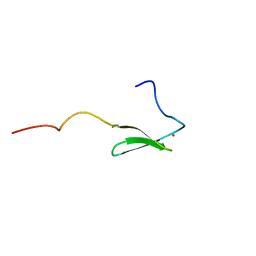 | | N-TERMINAL DOMAIN OF TFIIB, NMR | | Descriptor: | TFIIB, ZINC ION | | Authors: | Zhu, W, Zeng, Q, Colangelo, C.M, Lewis, L.M, Summers, M.F, Scott, R.A. | | Deposit date: | 1996-03-27 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon.
Nat.Struct.Biol., 3, 1996
|
|
6V8D
 
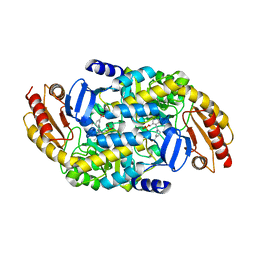 | | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators | | Descriptor: | (3Z)-3-iminocyclohex-1-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Zhu, W, Doubleday, P.T, Catlin, D.S, Weerawarna, P, Butrin, A, Shen, S, Kelleher, N.L, Liu, D, Silverman, R.B. | | Deposit date: | 2019-12-10 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators
To Be Published
|
|
5HI0
 
 | | The Substrate Binding Mode and Chemical Basis of a Reaction Specificity Switch in Oxalate Decarboxylase | | Descriptor: | COBALT (II) ION, OXALATE ION, Oxalate decarboxylase OxdC, ... | | Authors: | Zhu, W, Easthon, L.M, Reinhardt, L.A, Tu, C, Cohen, S.E, Silverman, D.N, Allen, K.N, Richards, N.G.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Substrate Binding Mode and Molecular Basis of a Specificity Switch in Oxalate Decarboxylase.
Biochemistry, 55, 2016
|
|
6V8C
 
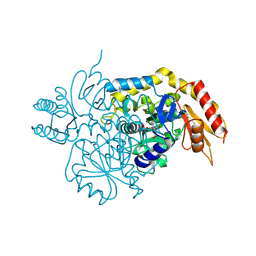 | | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators | | Descriptor: | 3-aminocyclohexa-1,3-diene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Zhu, W, Doubleday, P.T, Catlin, D.S, Weerawarna, P, Butrin, A, Shen, S, Kelleher, N.L, Liu, D, Silverman, R.B. | | Deposit date: | 2019-12-10 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators
To Be Published
|
|
4H2M
 
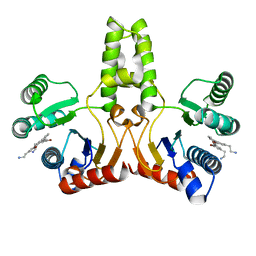 | | Structure of E. coli undecaprenyl diphosphate synthase in complex with BPH-1408 | | Descriptor: | 2,2'-{benzene-1,3-diylbis[ethyne-2,1-diyl(5-bromobenzene-3,1-diyl)]}diethanamine, Undecaprenyl pyrophosphate synthase | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2012-09-12 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Antibacterial drug leads targeting isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H3A
 
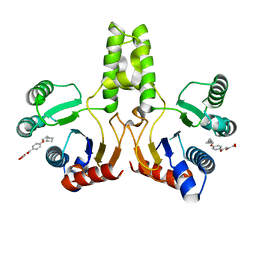 | |
4H2O
 
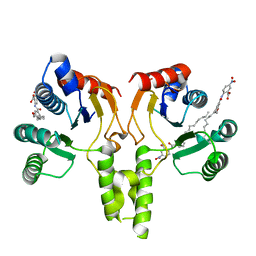 | |
4H2J
 
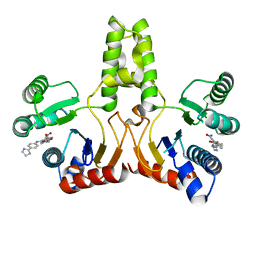 | |
4H8E
 
 | |
4H38
 
 | |
4H3C
 
 | |
6SCE
 
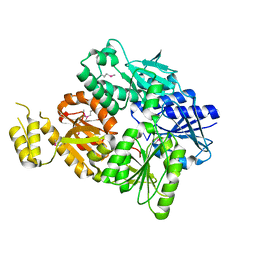 | | Structure of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Zhu, W, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate.
Nat Commun, 11, 2020
|
|
1J77
 
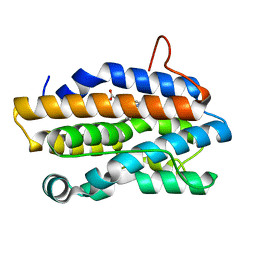 | | Crystal Structure of Gram-negative Bacterial Heme Oxygenase Complexed with Heme | | Descriptor: | HemO, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schuller, D.J, Zhu, W, Stojiljkovic, I, Wilks, A, Poulos, T.L. | | Deposit date: | 2001-05-15 | | Release date: | 2001-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of heme oxygenase from the gram-negative pathogen Neisseria meningitidis and a comparison with mammalian heme oxygenase-1.
Biochemistry, 40, 2001
|
|
4ET7
 
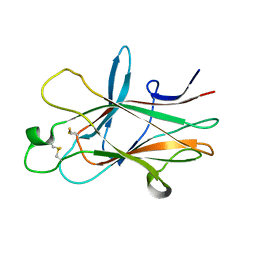 | |
3OLJ
 
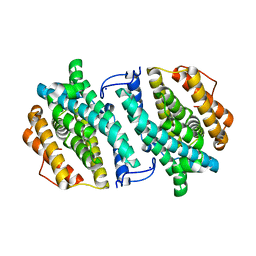 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) | | Descriptor: | Ribonucleoside-diphosphate reductase subunit M2, SODIUM ION | | Authors: | Chen, X.H, Xu, Z.J, Chen, B.E, Jiang, H.J, Yang, C.G, Zhu, W.L, Shao, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | hRRM2
To be Published
|
|
8BFS
 
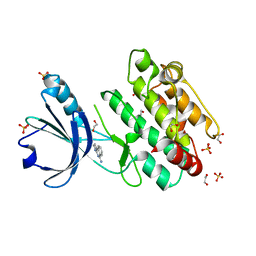 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326 | | Descriptor: | 1,2-ETHANEDIOL, 3~{H}-pyrrolo[2,3-c]isoquinolin-5-amine, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Zhu, W.F, Hernandez-Olmos, V, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326
To Be Published
|
|
3VPN
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
3VPM
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
3VPO
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
5ZBF
 
 | |
