5ZWJ
 
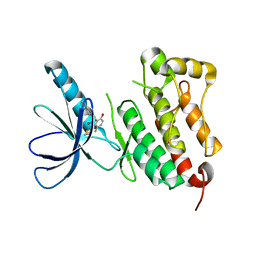 | | Crystal structure of EGFR 675-1022 T790M/C797S/V948R in complex with EAI045 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor | | Authors: | Zhao, P, Yun, C.H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of EGFR T790M/C797S/V948R in complex with EAI045.
Biochem. Biophys. Res. Commun., 502, 2018
|
|
7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
7VH5
 
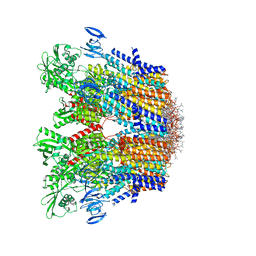 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the autoinhibited state (pH 7.4, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Plasma membrane ATPase 1, SPHINGOSINE | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
8JAT
 
 | |
6JTD
 
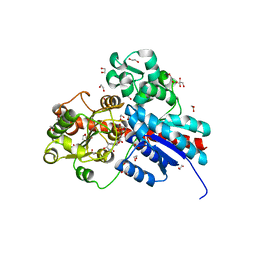 | | Crystal structure of TcCGT1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, C-glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhao, P, Yun, C.H. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular and Structural Characterization of a Promiscuous C-Glycosyltransferase from Trollius chinensis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
9BKJ
 
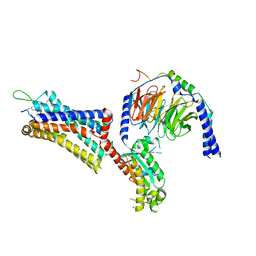 | | Cholecystokinin 1 receptor (CCK1R) Y140A mutant, Gq chimera (mGsqi) complex | | Descriptor: | AMINO GROUP, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Cary, B.P, Harikumar, K.G, Zhao, P, Desai, A.J, Mobbs, J.M, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling
To Be Published
|
|
9BKK
 
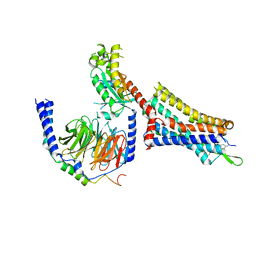 | | Cholecystokinin 1 receptor (CCK1R) sterol 7M mutant, Gq chimera (mGsqi) complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Harikumar, K.G, Zhao, P, Cary, B.P, Xu, X, Desai, A.J, Mobbs, J.I, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling
To Be Published
|
|
8JZN
 
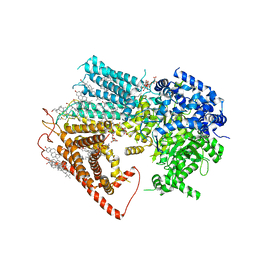 | | Structure of a fungal 1,3-beta-glucan synthase | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, C, You, Z, Chen, D, Hang, J, Wang, Z, Meng, J, Wang, L, Zhao, P, Qiao, J, Yun, C, Bai, L. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structure of a fungal 1,3-beta-glucan synthase.
Sci Adv, 9, 2023
|
|
6B3J
 
 | | 3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex | | Descriptor: | Exendin-P5, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Glukhova, A, Furness, S.G.B, Koole, C, Zhao, P, Clydesdale, L, Thal, D.M, Radjainia, M, Danev, R, Baumeister, W, Wang, M.W, Miller, L.J, Christopoulos, A, Sexton, P.M, Wootten, D. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-21 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex.
Nature, 555, 2018
|
|
3WVJ
 
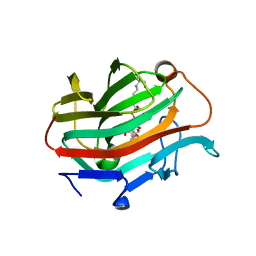 | | The crystal structure of native glycosidic hydrolase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase | | Authors: | Chen, C.C, Huang, J.W, Zhao, P, Ko, T.P, Huang, C.H, Chan, H.C, Huang, Z, Liu, W, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-05-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses and yeast production of the beta-1,3-1,4-glucanase catalytic module encoded by the licB gene of Clostridium thermocellum.
Enzyme.Microb.Technol., 71, 2015
|
|
8IUI
 
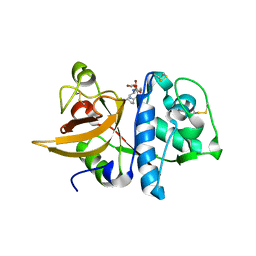 | |
4E8H
 
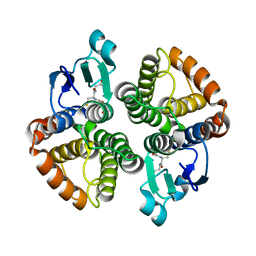 | | Structural of Bombyx mori glutathione transferase BmGSTD1 complex with GTT | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Tan, X, Ma, X.X, Hu, X.M, Chen, Q.M, Zhao, P, Xia, Q.Y, Zhou, C.Z. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural of Bombyx mori glutathione transferase BmGSTD1 complex with GTT
To be Published
|
|
4E8E
 
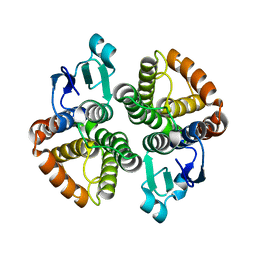 | | Structural characterization of Bombyx mori glutathione transferase BmGSTD1 | | Descriptor: | Glutathione S-transferase | | Authors: | Tan, X, Ma, X.X, Hu, X.M, Chen, Q.M, Zhao, P, Xia, Q.Y, Zhou, C.Z. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural characterization of Bombyx mori glutathione transferase BmGSTD1
To be Published
|
|
4R9I
 
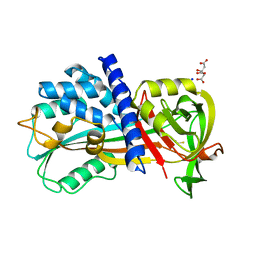 | | Crystal structure of cysteine proteinase inhibitor Serpin18 from Bombyx mori | | Descriptor: | BETA-MERCAPTOETHANOL, CITRATE ANION, SODIUM ION, ... | | Authors: | Guo, P.C, He, H.W, Zhao, P, Xia, Q.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the unique inhibitory mechanism of the silkworm protease inhibitor serpin18
Sci Rep, 5, 2015
|
|
5X2K
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with WZ4003 | | Descriptor: | Epidermal growth factor receptor, N-{3-[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Zhu, S.J, Zhao, P, Yun, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X5O
 
 | | Crystal structure of ZAK in complex with compound D2829 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, N-[2,4-bis(fluoranyl)-3-[2-(3-methoxy-1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]phenyl]-3-bromanyl-benzenesulfonamide | | Authors: | Dai, Y.B, Zhao, P, Yun, C.H. | | Deposit date: | 2017-02-17 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structure Based Design of N-(3-((1H-Pyrazolo[3,4-b]pyridin-5-yl)ethynyl)benzenesulfonamides as Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J. Med. Chem., 60, 2017
|
|
6KTH
 
 | | Crystal structure of Juvenile hormone diol kinase JHDK-L2 from silkworm, Bombyx mori | | Descriptor: | CALCIUM ION, GLYCEROL, Juvenile hormone diol kinase | | Authors: | Zhang, Y.S, Xu, H.Y, Wang, Z, Zhang, L, Zhao, P, Guo, P.C. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone diol kinase from the silkworm, Bombyx mori.
Int.J.Biol.Macromol., 167, 2021
|
|
