1DXS
 
 | |
8XZJ
 
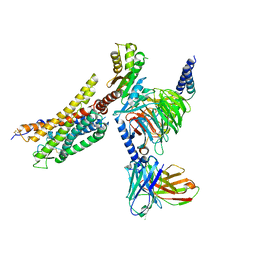 | | Cryo-EM structure of the WN353-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZG
 
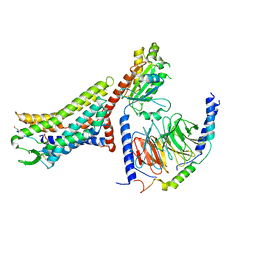 | | Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Apelin-13, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZH
 
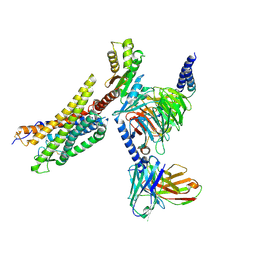 | | Cryo-EM structure of the MM07-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZF
 
 | | Cryo-EM structure of the WN561-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZI
 
 | | Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex | | Descriptor: | (3~{S})-5-methyl-3-[[1-pentan-3-yl-2-(thiophen-2-ylmethyl)benzimidazol-5-yl]carbonylamino]hexanoic acid, Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
1FO6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE | | Descriptor: | N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE, XENON | | Authors: | Wang, W.-C, Hsu, W.-H, Chien, F.-T, Chen, C.-Y. | | Deposit date: | 2000-08-25 | | Release date: | 2001-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and site-directed mutagenesis studies of N-carbamoyl-D-amino-acid amidohydrolase from Agrobacterium radiobacter reveals a homotetramer and insight into a catalytic cleft.
J.Mol.Biol., 306, 2001
|
|
4Y05
 
 | | KIF2C short Loop2 construct | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2C, MAGNESIUM ION, ... | | Authors: | Wang, W, Knossow, M, Gigant, B. | | Deposit date: | 2015-02-05 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | New Insights into the Coupling between Microtubule Depolymerization and ATP Hydrolysis by Kinesin-13 Protein Kif2C.
J.Biol.Chem., 290, 2015
|
|
1GSO
 
 | |
2DYU
 
 | |
2E2K
 
 | |
2E2L
 
 | |
2DYV
 
 | |
6IVE
 
 | |
5MIO
 
 | | KIF2C-DARPIN FUSION PROTEIN BOUND TO TUBULIN | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF2C,KIF2C FUSED TO A DARPIN,KIF2C FUSED TO A DARPIN, ... | | Authors: | Wang, W, Gigant, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin.
Nat Commun, 8, 2017
|
|
3CLH
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS)from Helicobacter pylori | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Wang, W.C, Liu, J.S, Cheng, W.C, Wang, H.J, Chen, Y.C. | | Deposit date: | 2008-03-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based inhibitor discovery of Helicobacter pylori dehydroquinate synthase.
Biochem.Biophys.Res.Commun., 373, 2008
|
|
3DBQ
 
 | | Crystal structure of TTK kinase domain | | Descriptor: | Dual specificity protein kinase TTK | | Authors: | Wang, W, Yang, Y.T, Gao, Y.F, Zhu, S.C, Wang, F, Old, W, Xu, Q.B, Resing, K, Ahn, N, Lei, M, Liu, X.D. | | Deposit date: | 2008-06-02 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Insights into Mps1 Kinase Activation
J.CELL.MOL.MED., 13, 2008
|
|
1R0M
 
 | | Structure of Deinococcus radiodurans N-acylamino acid racemase at 1.3 : insights into a flexible binding pocket and evolution of enzymatic activity | | Descriptor: | N-acylamino acid racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
2GGK
 
 | |
2GGJ
 
 | |
2GGL
 
 | |
2GGI
 
 | |
2GGH
 
 | |
2GGG
 
 | |
1XPY
 
 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-acylamino acid racemase, N~2~-ACETYL-L-GLUTAMINE | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-10 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
