4FXJ
 
 | |
4FXF
 
 | |
1S7Z
 
 | | Structure of Ocr from Bacteriophage T7 | | Descriptor: | CESIUM ION, Gene 0.3 protein | | Authors: | Walkinshaw, M.D, Taylor, P, Sturrock, S.S, Atanasiu, C, Berg, T, Henderson, R.M, Edwardson, J.M, Dryden, D.T. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of Ocr from Bacteriophage T7, a Protein that Mimics B-Form DNA
Mol.Cell, 9, 2002
|
|
3FWT
 
 | | Crystal structure of Leishmania major MIF2 | | Descriptor: | Macrophage migration inhibitory factor-like protein | | Authors: | Walkinshaw, M.D, Richardson, J.M. | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Leishmania major orthologues of macrophage migration inhibitory factor
Biochem.Biophys.Res.Commun., 380, 2009
|
|
3IS4
 
 | | Crystal structure of Leishmania mexicana pyruvate kinase (LmPYK)in complex with 1,3,6,8-pyrenetetrasulfonic acid | | Descriptor: | GLYCEROL, Pyruvate kinase, pyrene-1,3,6,8-tetrasulfonic acid | | Authors: | Walkinshaw, M.D, Morgan, H.P. | | Deposit date: | 2009-08-25 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An improved strategy for the crystallization of Leishmania mexicana pyruvate kinase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2F7T
 
 | | Crystal structure of the catalytic domain of Mos1 mariner transposase | | Descriptor: | MAGNESIUM ION, Mos1 transposase | | Authors: | Richardson, J.M, Dawson, A, Taylor, P, Finnegan, D.J, Walkinshaw, M.D. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Mos1 transposition: insights from structural analysis
Embo J., 25, 2006
|
|
3C2I
 
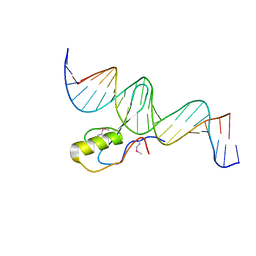 | | The Crystal Structure of Methyl-CpG Binding Domain of Human MeCP2 in Complex with a Methylated DNA Sequence from BDNF | | Descriptor: | DNA (5'-D(*DAP*DTP*DAP*DGP*DAP*DAP*DGP*DAP*DAP*DTP*DTP*DCP*(5CM)P*DGP*DTP*DTP*DCP*DCP*DAP*DG)-3'), DNA (5'-D(*DTP*DCP*DTP*DGP*DGP*DAP*DAP*(5CM)P*DGP*DGP*DAP*DAP*DTP*DTP*DCP*DTP*DTP*DCP*DTP*DA)-3'), Methyl-CpG-binding protein 2 | | Authors: | Ho, K.L, McNae, I.W, Schmiedeberg, L, Klose, R.J, Bird, A.P, Walkinshaw, M.D. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MeCP2 binding to DNA depends upon hydration at methyl-CpG
Mol.Cell, 29, 2008
|
|
1LJ1
 
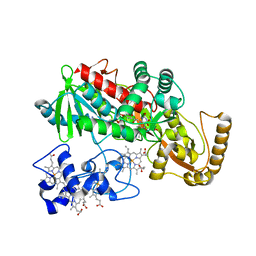 | | Crystal structure of Q363F/R402A mutant flavocytochrome c3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mowat, C.G, Pankhurst, K.L, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-04-18 | | Release date: | 2002-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering Water to act as the active site acid catalyst in a soluble fumarate reductase
Biochemistry, 41, 2002
|
|
1M64
 
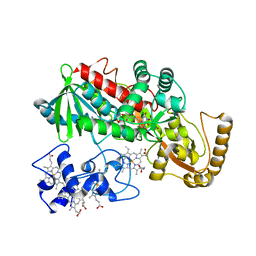 | | Crystal structure of Q363F mutant flavocytochrome c3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mowat, C.G, Pankhurst, K.L, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-07-12 | | Release date: | 2002-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering water to act as an active site acid catalyst in a soluble fumarate reductase
Biochemistry, 41, 2002
|
|
3KTX
 
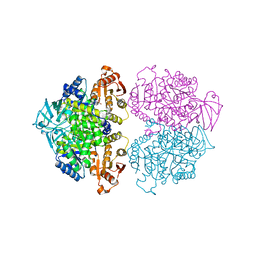 | | Crystal structure of Leishmania mexicana pyruvate kinase (LmPYK)in complex with 1,3,6,8-pyrenetetrasulfonic acid | | Descriptor: | GLYCEROL, Pyruvate kinase, pyrene-1,3,6,8-tetrasulfonic acid | | Authors: | Morgan, H.P, Walkinshaw, M.D. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An improved strategy for the crystallization of Leishmania mexicana pyruvate kinase.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2QIJ
 
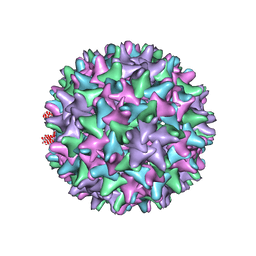 | | Hepatitis B Capsid Protein with an N-terminal extension modelled into 8.9 A data. | | Descriptor: | Core antigen | | Authors: | Tan, W.S, McNae, I.W, Ho, K.L, Walkinshaw, M.D. | | Deposit date: | 2007-07-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (8.9 Å) | | Cite: | Crystallization and X-ray analysis of the T = 4 particle of hepatitis B capsid protein with an N-terminal extension.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1W8V
 
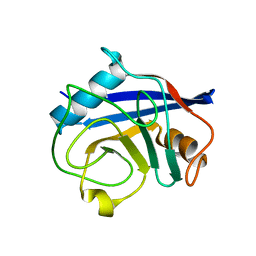 | |
1W8M
 
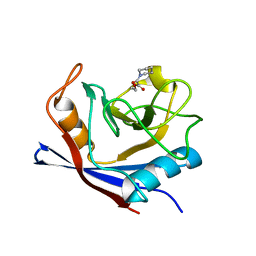 | |
4WJ8
 
 | | Human Pyruvate Kinase M2 Mutant C424A | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mitchell, T, Yuan, M, McNae, I, Morgan, H, Walkinshaw, M.D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Human Pyruvate Kinase M2 Mutant C424A
To Be Published
|
|
2P32
 
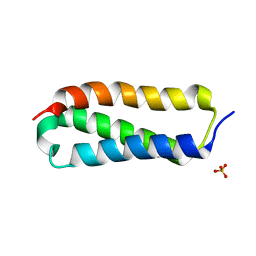 | |
5NOW
 
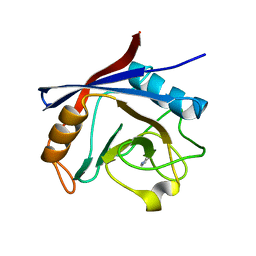 | | Structure of cyclophilin A in complex with pyridine-3,4-diamine | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, pyridine-3,4-diamine | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOV
 
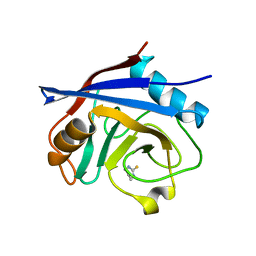 | | Structure of cyclophilin A in complex with hexahydropyrimidine-2-thione | | Descriptor: | 1,3-diazinane-2-thione, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOZ
 
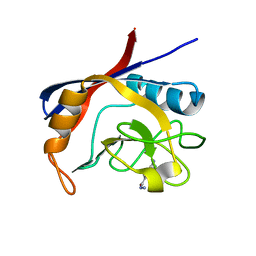 | | Structure of cyclophilin A in complex with 3,4-diaminobenzohydrazide | | Descriptor: | 3,4-bis(azanyl)benzohydrazide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
1A58
 
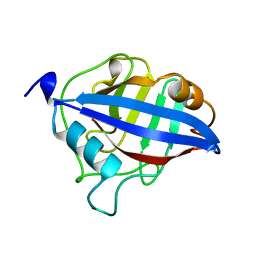 | | CYCLOPHILIN FROM BRUGIA MALAYI | | Descriptor: | CYCLOPHILIN | | Authors: | Taylor, P, Page, A.P, Kontopidis, G, Husi, H, Walkinshaw, M.D. | | Deposit date: | 1998-02-20 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The X-ray structure of a divergent cyclophilin from the nematode parasite Brugia malayi.
FEBS Lett., 425, 1998
|
|
1BCK
 
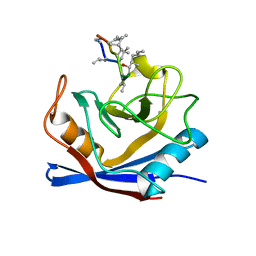 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 2-THR CYCLOSPORIN | | Descriptor: | CYCLOSPORIN C, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-04-30 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
5LUD
 
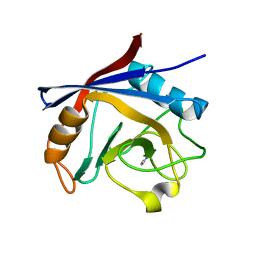 | | Structure of Cyclophilin A in complex with 2,3-Diaminopyridine | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, pyridine-2,3-diamine | | Authors: | McNae, I.W, Nowicki, M.W, Blackburn, E.A, Wear, M.A, Walkinshaw, M.D. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermo-kinetic analysis space expansion for cyclophilin-ligand interactions - identification of a new nonpeptide inhibitor using BiacoreTM T200.
FEBS Open Bio, 7, 2017
|
|
5EIR
 
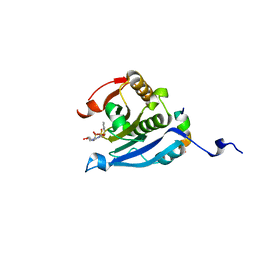 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E, SULFATE ION, ... | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
5EI3
 
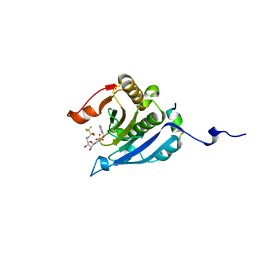 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | Eukaryotic translation initiation factor 4 gamma, Eukaryotic translation initiation factor 4E, SULFATE ION, ... | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-29 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
5EKV
 
 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | 3-[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]-4-oxidanyl-cyclobut-3-ene-1,2-dione, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
5EHC
 
 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | 3-[[(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-7-[(3-chlorophenyl)methyl]-6-oxidanylidene-1~{H}-purin-7-ium-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]-4-oxidanyl-cyclobut-3-ene-1,2-dione, Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
