8WFJ
 
 | |
8WFK
 
 | |
8WFL
 
 | |
8WFI
 
 | |
5I8J
 
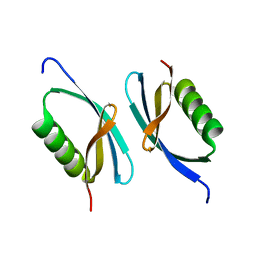 | | Crystal structure of Dmd from phage RB69 | | Descriptor: | Dmd discriminator of mRNA degradation | | Authors: | Wei, Y, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterizations of phage antitoxin Dmd and its interactions with bacterial toxin RnlA
Biochem. Biophys. Res. Commun., 472, 2016
|
|
6P6B
 
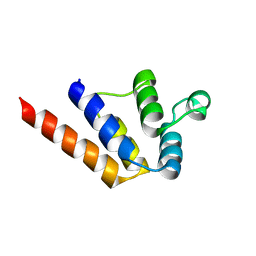 | |
6P6C
 
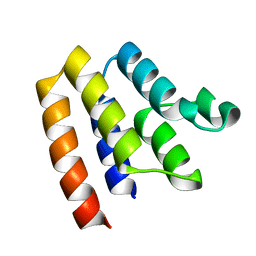 | |
2PP4
 
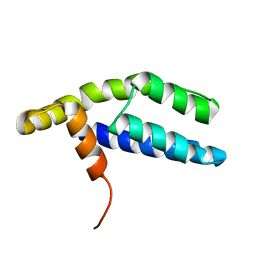 | | Solution Structure of ETO-TAFH refined in explicit solvent | | Descriptor: | Protein ETO | | Authors: | Wei, Y, Liu, S, Lausen, J, Woodrell, C, Cho, S, Biris, N, Kobayashi, N, Yokoyama, S, Werner, M.H. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription
Nat.Struct.Mol.Biol., 14, 2007
|
|
1ESC
 
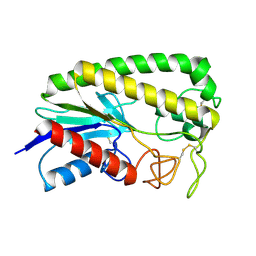 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
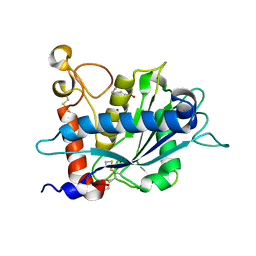
3B92
 
 | |
4EFE
 
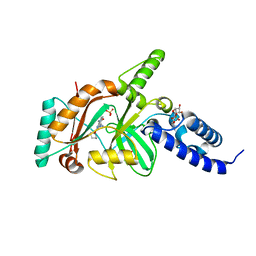 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
4EFB
 
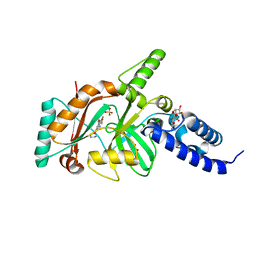 | | Crystal structure of DNA ligase | | Descriptor: | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DNA ligase
To be Published
|
|
2OIN
 
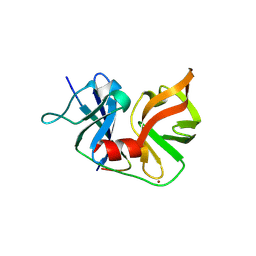 | | crystal structure of HCV NS3-4A R155K mutant | | Descriptor: | NS4A peptide, Polyprotein, ZINC ION | | Authors: | Wei, Y. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha.
J.Biol.Chem., 282, 2007
|
|
1F1J
 
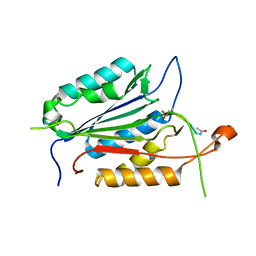 | |
1FTN
 
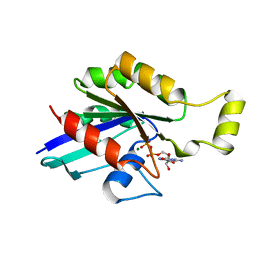 | | CRYSTAL STRUCTURE OF THE HUMAN RHOA/GDP COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN RHOA (H12) | | Authors: | Wei, Y, Zhang, Y, Derewenda, U, Liu, X, Minor, W, Nakamoto, R.K, Somlyo, A.V, Somlyo, A.P, Derewenda, Z.S. | | Deposit date: | 1997-03-13 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RhoA-GDP and its functional implications.
Nat.Struct.Biol., 4, 1997
|
|
1JKM
 
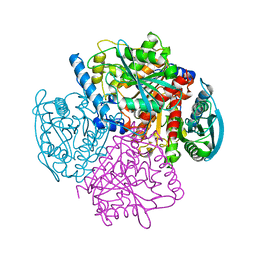 | | BREFELDIN A ESTERASE, A BACTERIAL HOMOLOGUE OF HUMAN HORMONE SENSITIVE LIPASE | | Descriptor: | BREFELDIN A ESTERASE | | Authors: | Wei, Y, Contreras, A.J, Sheffield, P, Osterlund, T, Derewenda, U, Matern, U.O, Derewenda, Z.S. | | Deposit date: | 1998-02-04 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase.
Nat.Struct.Biol., 6, 1999
|
|
3G7E
 
 | |
4I8O
 
 | |
2OI0
 
 | | Crystal structure analysis 0f the TNF-a Coverting Enzyme (TACE) in complexed with Aryl-sulfonamide | | Descriptor: | (3S)-1-{[4-(BUT-2-YN-1-YLOXY)PHENYL]SULFONYL}PYRROLIDINE-3-THIOL, TNF- a Converting Enzyme (TACE), ZINC ION | | Authors: | Wei, Y, Rao, G.B, Bandarage, U.K. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel thiol-based TACE inhibitors: rational design, synthesis, and SAR of thiol-containing aryl sulfonamides
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1ESE
 
 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | DIETHYL PHOSPHONATE, ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
1ESD
 
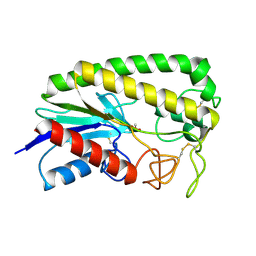 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | ESTERASE, METHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
8HMB
 
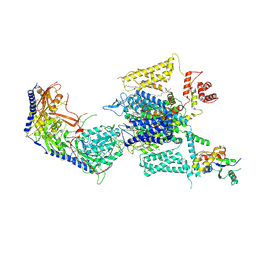 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with benidipine (BEN) | | Descriptor: | (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-4-(3-nitrophenyl)piperidine-3,5-dicarboxylate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMA
 
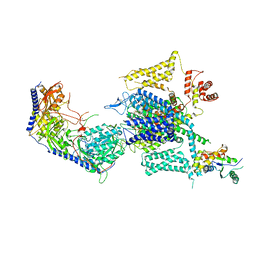 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HLP
 
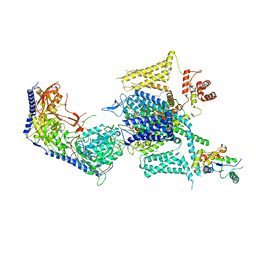 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
2QV1
 
 | | Crystal structure of HCV NS3-4A V36M mutant | | Descriptor: | NS3, ZINC ION, peptide | | Authors: | Wei, Y. | | Deposit date: | 2007-08-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phenotypic and Structural Analyses of HCV NS3 Protease Val36 Variants
To be Published
|
|
