8HFB
 
 | | Evolved variant of quercetin 2,4-dioxygenase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Eom, H, Song, W.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Underlying Role of Hydrophobic Environments in Tuning Metal Elements for Efficient Enzyme Catalysis.
J.Am.Chem.Soc., 145, 2023
|
|
2L53
 
 | |
5C2V
 
 | | Kuenenia stuttgartiensis Hydrazine Synthase | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEME C, ... | | Authors: | Dietl, A, Ferousi, C, Maalcke, W.J, Menzel, A, de Vries, S, Keltjens, J.T, Jetten, M.S.M, Kartal, B, Barends, T.R.M. | | Deposit date: | 2015-06-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The inner workings of the hydrazine synthase multiprotein complex.
Nature, 527, 2015
|
|
2LJR
 
 | | GLUTATHIONE TRANSFERASE APO-FORM FROM HUMAN | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
2M0X
 
 | |
2MB3
 
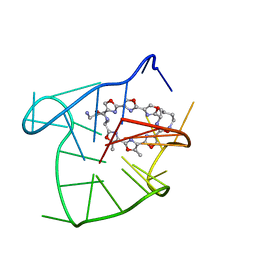 | | Solution structure of an intramolecular (3+1) human telomeric G-quadruplex bound to a telomestatin derivative | | Descriptor: | (12S,27S)-12,27-bis(4-aminobutyl)-4,30-dimethyl-3,7,14,18,22,29-hexaoxa-11,26,31,32,33,34,35,36-octaazaheptacyclo[26.2. 1.1~2,5~.1~6,9~.1~13,16~.1~17,20~.1~21,24~]hexatriaconta-1(30),2(36),4,6(35),8,13(34),15,17(33),19,21(32),23,28(31)-dode caene-10,25-dione, DNA_(5'-D(*TP*TP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*A)-3') | | Authors: | Chung, W.J, Heddi, B, Tera, M, Iida, K, Nagasawa, K, Phan, A.T. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular (3 + 1) human telomeric g-quadruplex bound to a telomestatin derivative.
J.Am.Chem.Soc., 135, 2013
|
|
5DLV
 
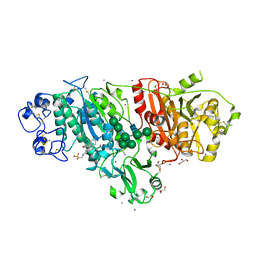 | | Crystal structure of Autotaxin (ENPP2) with tauroursodeoxycholic acid (TUDCA) | | Descriptor: | 2-{[(3alpha,5beta,7alpha,8alpha,14beta,17alpha)-3,7-dihydroxy-24-oxocholan-24-yl]amino}ethanesulfonic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.J, Heidebrecht, T, von Castelmur, E, Joosten, R.P, Perrakis, A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid binding to Autotaxin links bile salts and lysophosphatidic acid signalling.
Nat Commun, 7, 2016
|
|
5DU3
 
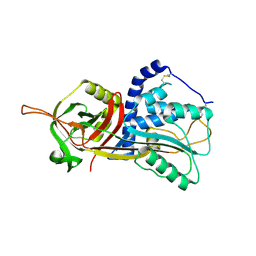 | | Active form of human C1-inhibitor | | Descriptor: | Plasma protease C1 inhibitor | | Authors: | Pannu, N.S, Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A. | | Deposit date: | 2015-09-18 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
2M4N
 
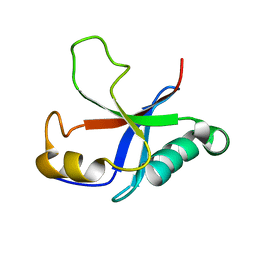 | | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans | | Descriptor: | Protein AFD-1, isoform a | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Seidel, R.D, Liddington, R.C, Weis, W.I, Nelson, W.J, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans
To be Published
|
|
1AVP
 
 | |
5DBR
 
 | |
2N3M
 
 | |
5DQO
 
 | |
2O2F
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-(4-BENZYL-4-METHOXYPIPERIDIN-1-YL)-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
1D1O
 
 | |
2O21
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2O2N
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-[4-(BIPHENYL-2-YLMETHYL)PIPERAZIN-1-YL]-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2O22
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | Apoptosis regulator Bcl-2, N-[(4-{[1,1-dimethyl-2-(phenylthio)ethyl]amino}-3-nitrophenyl)sulfonyl]-4-(4,4-dimethylpiperidin-1-yl)benzamide | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
5DUQ
 
 | | Active human c1-inhibitor in complex with dextran sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasma protease C1 inhibitor, SULFITE ION, ... | | Authors: | Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A, Pannu, N.S. | | Deposit date: | 2015-09-20 | | Release date: | 2016-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
2P7V
 
 | | Crystal structure of the Escherichia coli regulator of sigma 70, Rsd, in complex with sigma 70 domain 4 | | Descriptor: | MAGNESIUM ION, RNA polymerase sigma factor rpoD, Regulator of sigma D | | Authors: | Patikoglou, G.A, Westblade, L.F, Campbell, E.A, Lamour, V, Lane, W.J, Darst, S.A. | | Deposit date: | 2007-03-20 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4.
J.Mol.Biol., 372, 2007
|
|
5EMV
 
 | | Crystal structure of the palmitoylated human TEAD2 transcription factor | | Descriptor: | Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Gierke, S, Schnier, P.D, Murray, J, Sandoval, W.N, Sagolla, M, Dey, A, Hannoush, R.N, Fairbrother, W.J, Cunningham, C.N. | | Deposit date: | 2015-11-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling.
Structure, 24, 2016
|
|
2O1Y
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2NSV
 
 | |
2NSW
 
 | |
5EMW
 
 | | Crystal structure of the palmitoylated human TEAD3 transcription factor | | Descriptor: | CALCIUM ION, Transcriptional enhancer factor TEF-5 | | Authors: | Noland, C.L, Gierke, S, Schnier, P.D, Murray, J, Sandoval, W.N, Sagolla, M, Dey, A, Hannoush, R.N, Fairbrother, W.J, Cunningham, C.N. | | Deposit date: | 2015-11-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling.
Structure, 24, 2016
|
|
