4XQ8
 
 | |
1D9S
 
 | | TUMOR SUPPRESSOR P15(INK4B) STRUCTURE BY COMPARATIVE MODELING AND NMR DATA | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR B | | Authors: | Yuan, C, Ji, L, Selby, T.L, Byeon, I.J.L, Tsai, M.D. | | Deposit date: | 1999-10-29 | | Release date: | 2000-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: comparisons of conformational properties between p16(INK4A) and p18(INK4C).
J.Mol.Biol., 294, 1999
|
|
8XMI
 
 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-12-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis and synergism of ATP and Na + activation in bacterial K + uptake system KtrAB.
Nat Commun, 15, 2024
|
|
8XMH
 
 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-12-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis and synergism of ATP and Na + activation in bacterial K + uptake system KtrAB.
Nat Commun, 15, 2024
|
|
2BAX
 
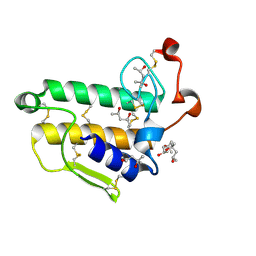 | | Atomic Resolution Structure of the Double Mutant (K53,56M) of Bovine Pancreatic Phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Dauter, Z, Dauter, M, Tsai, M.D. | | Deposit date: | 2005-10-15 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2B96
 
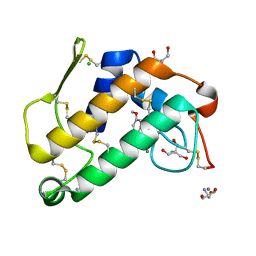 | | Third Calcium ion found in an inhibitor bound phospholipase A2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-METHOXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Yamane, T, Tsai, M.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Third Calcium ion found in an inhibitor bound phospholipase A2
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4XRH
 
 | |
5XZW
 
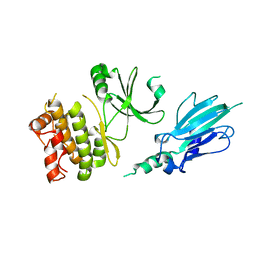 | | Crystal structure of Rad53 1-466 | | Descriptor: | Serine/threonine-protein kinase RAD53 | | Authors: | Weng, J.H, Tsai, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phospho-Priming Confers Functionally Relevant Specificities for Rad53 Kinase Autophosphorylation
Biochemistry, 56, 2017
|
|
6ISI
 
 | | Structure of 9N-I DNA polymerase incorporation with 3'-CL in the active site | | Descriptor: | 3-[2-[2-(2-azanylethoxy)ethoxy]ethoxy]propanoic acid, CALCIUM ION, DNA (5'-D(P*AP*CP*GP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6ISG
 
 | | Structure of 9N-I DNA polymerase incorporation with dG in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*CP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6ISF
 
 | | Structure of 9N-I DNA polymerase incorporation with dT in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*T)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6ISH
 
 | | Structure of 9N-I DNA polymerase incorporation with 3'-AL in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*(DAL))-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6IS7
 
 | | Structure of 9N-I DNA polymerase incorporation with dA in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*A)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
5DDY
 
 | | Binary complex of human Polymerase lambda with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase lambda, MAGNESIUM ION, ... | | Authors: | Liu, M.S, Tsai, M.D. | | Deposit date: | 2015-08-25 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural Mechanism for the Fidelity Modulation of DNA Polymerase lambda
J.Am.Chem.Soc., 138, 2016
|
|
5DKW
 
 | |
1C74
 
 | | Structure of the double mutant (K53,56M) of phospholipase A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Tsai, M.D, Jain, M.K, Ramakumar, S. | | Deposit date: | 2000-01-22 | | Release date: | 2000-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the anionic interface preference and k*cat activation of pancreatic phospholipase A2.
Biochemistry, 39, 2000
|
|
1GH4
 
 | | Structure of the triple mutant (K56M, K120M, K121M) of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Velmurugan, D, Tsai, M.D. | | Deposit date: | 2000-11-09 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Observation of additional calcium ion in the crystal structure of the triple mutant K56,120,121M of bovine pancreatic phospholipase A2.
J.Mol.Biol., 324, 2002
|
|
5XZV
 
 | | Crystal structure of Rad53 1-466 in complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase RAD53 | | Authors: | Weng, J.H, Tsai, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phospho-Priming Confers Functionally Relevant Specificities for Rad53 Kinase Autophosphorylation
Biochemistry, 56, 2017
|
|
5ZLC
 
 | | Binary complex of human DNA Polymerase Mu with MndGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA-directed DNA/RNA polymerase mu, MANGANESE (II) ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-03-27 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6A33
 
 | | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA | | Descriptor: | 15-mer peptide from TRAF-interacting protein with FHA domain-containing protein A, TNF receptor-associated factor 6 | | Authors: | Huang, W.C, Liao, J.H, Hsiao, T.C, Maestre-Reyna, M, Bessho, Y, Tsai, M.D. | | Deposit date: | 2018-06-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA.
Chembiochem, 20, 2019
|
|
2AFF
 
 | | The solution structure of the Ki67FHA/hNIFK(226-269)3P complex | | Descriptor: | Antigen KI-67, MKI67 FHA domain interacting nucleolar phosphoprotein | | Authors: | Byeon, I.-J.L, Li, H, Song, H, Gronenborn, A.M, Tsai, M.D. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67.
Nat.Struct.Mol.Biol., 12, 2005
|
|
8KE3
 
 | | PylRS C-terminus domain mutant bound with D-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89980984 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE2
 
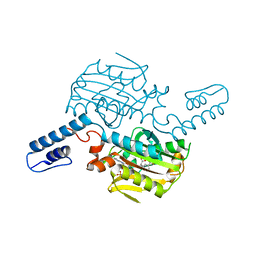 | | PylRS C-terminus domain mutant bound with L-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2S)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19639969 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE5
 
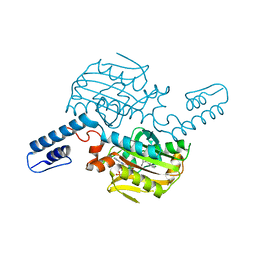 | | PylRS C-terminus domain mutant bound with D-3-chlorophenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(3-chlorophenyl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.900073 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE1
 
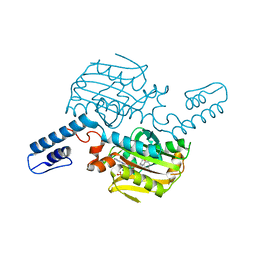 | | PylRS C-terminus domain mutant bound with L-3-bromophenylalanine and AMPNP | | Descriptor: | 3-bromo-L-phenylalanine, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.50081539 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
