2BDI
 
 | | Human Kallikrein 4 complex with cobalt and p-aminobenzamidine | | Descriptor: | COBALT (II) ION, Kallikrein-4, P-AMINO BENZAMIDINE | | Authors: | Debela, M, Bode, W, Goettig, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site.
J.Mol.Biol., 362, 2006
|
|
2BDH
 
 | | Human Kallikrein 4 complex with zinc and p-aminobenzamidine | | Descriptor: | Kallikrein-4, P-AMINO BENZAMIDINE, ZINC ION | | Authors: | Debela, M, Bode, W, Goettig, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site.
J.Mol.Biol., 362, 2006
|
|
2BDG
 
 | | Human Kallikrein 4 complex with nickel and p-aminobenzamidine | | Descriptor: | CHLORIDE ION, Kallikrein-4, NICKEL (II) ION, ... | | Authors: | Debela, M, Bode, W, Goettig, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site.
J.Mol.Biol., 362, 2006
|
|
2D30
 
 | | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution | | Descriptor: | ZINC ION, cytidine deaminase | | Authors: | Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Moroz, O.V, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-09-21 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution
To be Published
|
|
1YUU
 
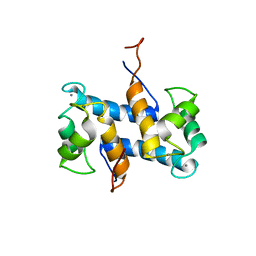 | | Solution structure of Calcium-S100A13 | | Descriptor: | CALCIUM ION, S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2A1Y
 
 | | Crystal Structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A Resolution. | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A resolution.
To be Published
|
|
1TXE
 
 | | Solution structure of the active-centre mutant Ile14Ala of the histidine-containing phosphocarrier protein (HPr) from Staphylococcus carnosus | | Descriptor: | Phosphocarrier protein HPr | | Authors: | Moeglich, A, Koch, B, Hengstenberg, W, Brunner, E, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-07-04 | | Release date: | 2005-03-08 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the active-centre mutant I14A of the histidine-containing phosphocarrier protein from Staphylococcus carnosus
Eur.J.Biochem., 271, 2004
|
|
1SW8
 
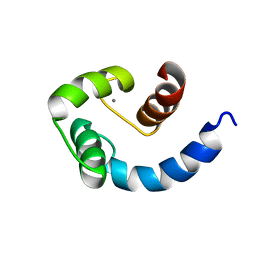 | | Solution structure of the N-terminal domain of Human N60D calmodulin refined with paramagnetism based strategy | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Bertini, I, Del Bianco, C, Gelis, I, Katsaros, N, Luchinat, C, Parigi, G, Peana, M, Provenzani, A, Zoroddu, M.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-30 | | Release date: | 2004-04-06 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Experimentally exploring the conformational space sampled by domain reorientation in calmodulin
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1R20
 
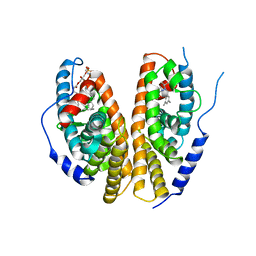 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI06830 | | Descriptor: | ECDYSONE RECEPTOR, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, N-(TERT-BUTYL)-3,5-DIMETHYL-N'-[(5-METHYL-2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)CARBONYL]BENZOHYDRAZIDE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-25 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
1R1K
 
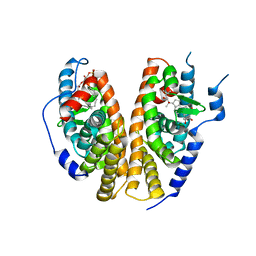 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.-M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-24 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
1RK9
 
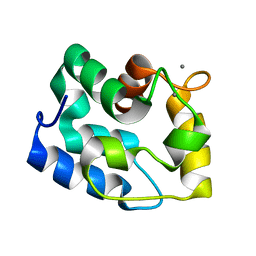 | | Solution Structure of Human alpha-Parvalbumin (Minimized Average Structure) | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Baig, I, Bertini, I, Del Bianco, C, Gupta, Y.K, Lee, Y.-M, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based refinement strategy for the solution structure of human alpha-parvalbumin
Biochemistry, 43, 2004
|
|
1RJV
 
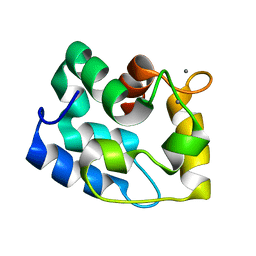 | | Solution Structure of Human alpha-Parvalbumin refined with a paramagnetism-based strategy | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Baig, I, Bertini, I, Del Bianco, C, Gupta, Y.K, Lee, Y.M, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-11-20 | | Release date: | 2004-05-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-Based Refinement Strategy for the Solution Structure of Human alpha-Parvalbumin.
Biochemistry, 43, 2004
|
|
1QZZ
 
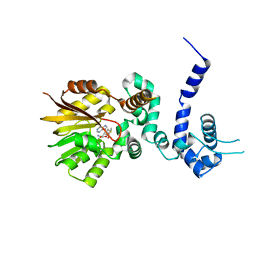 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
1SB6
 
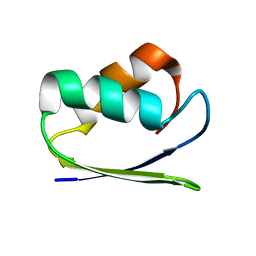 | | Solution structure of a cyanobacterial copper metallochaperone, ScAtx1 | | Descriptor: | copper chaperone ScAtx1 | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.C, Borrelly, G.P, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-02-10 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a Cyanobacterial Metallochaperone: INSIGHT INTO AN ATYPICAL COPPER-BINDING MOTIF.
J.Biol.Chem., 279, 2004
|
|
1S6U
 
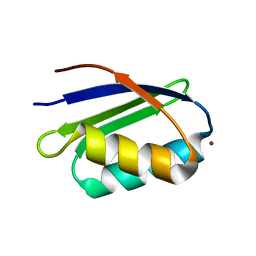 | | Solution structure and backbone dynamics of the Cu(I) form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-27 | | Release date: | 2004-04-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1S6O
 
 | | Solution structure and backbone dynamics of the apo-form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-26 | | Release date: | 2004-04-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1SZW
 
 | |
1SU3
 
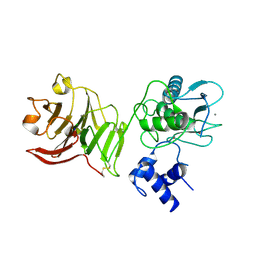 | | X-ray structure of human proMMP-1: New insights into collagenase action | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jozic, D, Bourenkov, G, Lim, N.H, Nagase, H, Bode, W, Maskos, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-26 | | Release date: | 2004-12-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of human proMMP-1: new insights into procollagenase activation and collagen binding.
J.Biol.Chem., 280, 2005
|
|
1TL5
 
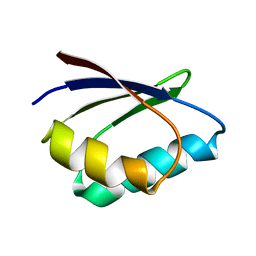 | | Solution structure of apoHAH1 | | Descriptor: | Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
1TW2
 
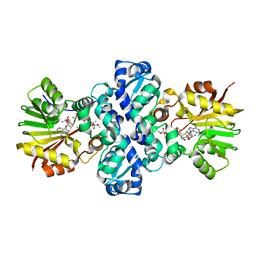 | | Crystal structure of Carminomycin-4-O-methyltransferase (DnrK) in complex with S-adenosyl-L-homocystein (SAH) and 4-methoxy-e-rhodomycin T (M-ET) | | Descriptor: | Carminomycin 4-O-methyltransferase, METHYL (4R)-2-ETHYL-2,5,12-TRIHYDROXY-7-METHOXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-RIBO-HEXOPYRANOSYL]OXY}-1H,2H,3H,4H,6H,11H-TETRACENE-1-CARBOXYLATE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jansson, A, Koskiniemi, H, Mantsala, P, Niemi, J, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products
J.Biol.Chem., 279, 2004
|
|
1Q7X
 
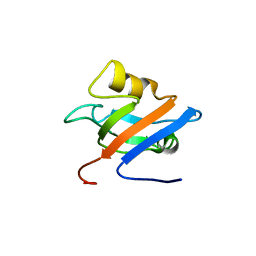 | | Solution structure of the alternatively spliced PDZ2 domain (PDZ2b) of PTP-Bas (hPTP1E) | | Descriptor: | PDZ2b domain of PTP-Bas (hPTP1E) | | Authors: | Kachel, N, Erdmann, K.S, Kremer, W, Wolff, P, Gronwald, W, Heumann, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-08-20 | | Release date: | 2003-12-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): Splicing induced modulation of ligand specificity.
J.Mol.Biol., 334, 2003
|
|
1TL4
 
 | | Solution structure of Cu(I) HAH1 | | Descriptor: | COPPER (I) ION, Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
1SO9
 
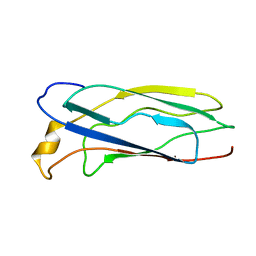 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1SJW
 
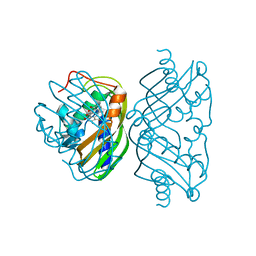 | | Structure of polyketide cyclase SnoaL | | Descriptor: | METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, nogalonic acid methyl ester cyclase | | Authors: | Sultana, A, Kallio, P, Jansson, A, Wang, J.S, Neimi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation.
Embo J., 23, 2004
|
|
1TTX
 
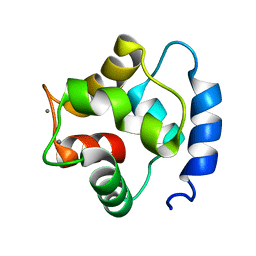 | | Solution Structure of human beta parvalbumin (oncomodulin) refined with a paramagnetism based strategy | | Descriptor: | CALCIUM ION, Oncomodulin | | Authors: | Babini, E, Bertini, I, Capozzi, F, Del Bianco, C, Hollender, D, Kiss, T, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-18 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human beta-Parvalbumin and Structural Comparison with Its Paralog alpha-Parvalbumin and with Their Rat Orthologs(,)
Biochemistry, 43, 2004
|
|
