1XK5
 
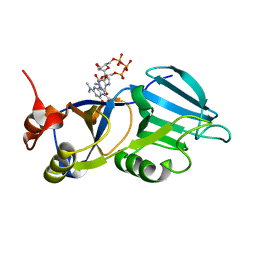 | | Crystal structure of the m3G-cap-binding domain of snurportin1 in complex with a m3GpppG-cap dinucleotide | | Descriptor: | 2,2,7-TRIMETHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, snurportin-1 | | Authors: | Strasser, A, Dickmanns, A, Luehrmann, R, Ficner, R. | | Deposit date: | 2004-09-27 | | Release date: | 2005-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for m(3)G-cap-mediated nuclear import of spliceosomal UsnRNPs by snurportin1
Embo J., 24, 2005
|
|
2VCG
 
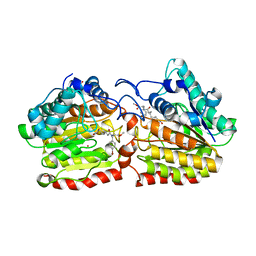 | | Crystal structure of a HDAC-like protein HDAH from Bordetella sp. with the bound inhibitor ST-17 | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Dickmanns, A, Strasser, A, Ficner, R. | | Deposit date: | 2007-09-24 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phenylalanine-Containing Hydroxamic Acids as Selective Inhibitors of Class Iib Histone Deacetylases (Hdacs).
Bioorg.Med.Chem., 16, 2008
|
|
3QBR
 
 | | BakBH3 in complex with sjA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Bcl-2 homologous antagonist/killer, SJCHGC06286 protein | | Authors: | Lee, E.F, Clarke, O.B, Fairlie, W.D, Colman, P.M, Evangelista, M, Feng, Z, Speed, T.P, Tchoubrieva, E, Strasser, A, Kalinna, B. | | Deposit date: | 2011-01-13 | | Release date: | 2011-04-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Discovery and molecular characterization of a Bcl-2-regulated cell death pathway in schistosomes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2QNA
 
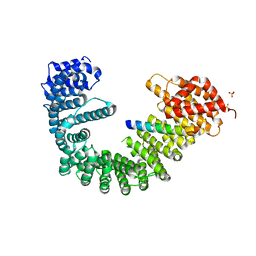 | | Crystal structure of human Importin-beta (127-876) in complex with the IBB-domain of Snurportin1 (1-65) | | Descriptor: | Importin subunit beta-1, SULFATE ION, Snurportin-1 | | Authors: | Wohlwend, D, Strasser, A, Dickmanns, A, Ficner, R. | | Deposit date: | 2007-07-18 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus.
J.Mol.Biol., 374, 2007
|
|
1RE6
 
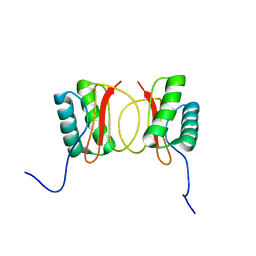 | | Localisation of Dynein Light Chains 1 and 2 and their Pro-apoptotic Ligands | | Descriptor: | dynein light chain 2 | | Authors: | Day, C.L, Puthalakath, H, Skea, G, Strasser, A, Barsukov, I, Lian, L.Y, Huang, D.C, Hinds, M.G. | | Deposit date: | 2003-11-06 | | Release date: | 2004-03-23 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Localization of dynein light chains 1 and 2 and their pro-apoptotic ligands.
Biochem.J., 377, 2004
|
|
